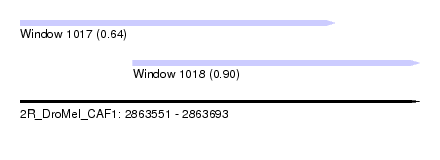
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,863,551 – 2,863,693 |
| Length | 142 |
| Max. P | 0.898927 |

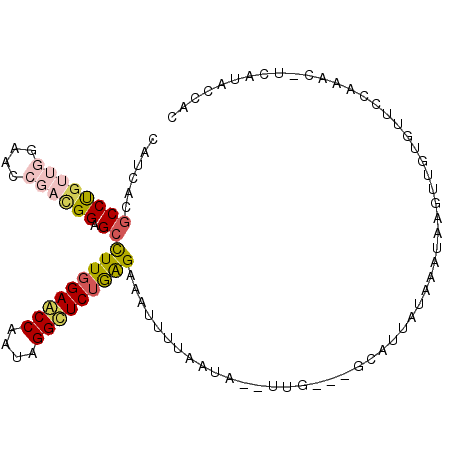
| Location | 2,863,551 – 2,863,663 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.75 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -19.26 |
| Energy contribution | -18.97 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.637786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2863551 112 + 20766785 UGGCAAAAGUGGAGAGCAGUGGAACCUGUUGUUGAAAGACCAUCACGCCUGUUGGAACCGACGGAGCCUUGGAACCAAUAGGUUCUGGGAAAUUUUAAUAAGUUG---GCAUUCU ((.(((..((((..(((((......)))))(((....)))..))))((((((((....)))))).))((..(((((....)))))..)).............)))---.)).... ( -32.90) >DroVir_CAF1 15051 108 + 1 UCGCAAAGCUGAAGCCCAAUGGAACAUGUUGCUGGAAUGUCUGCACGCCCGUGGGCACAGCCGGCGCCUUUGAACCAAUAGGCUCUAAGAAAAU-AAAGC--UU--AUAAAU--U .....(((((.........((((.......(((((..((((..(......)..))))...)))))((((.(......).)))))))).......-..)))--))--......--. ( -28.27) >DroSec_CAF1 15184 112 + 1 UGGCAAAAGUGGAUAGCAGUGGAACCUGUUGUUGAAAGACCAUCACGCCUGUUGGAACCGACGGAGCCUUGGAACCAAUAGGUUCUGGGAAAUUUUAAUAGGUUG---GCUUUAU .....(((((.((((((((......))))))))....((((.....((((((((....)))))).))((..(((((....)))))..))...........)))).---))))).. ( -36.10) >DroSim_CAF1 22055 112 + 1 UGGCAAAAGUGGAGAGCAGUGGAACCUGUUGUUGAAAGACCAUCACGCCUGUUGGAACCGAUGGAGCCUUGGAACCAAUAGGUUCUGGGAAAUUUUAAUAGGUUG---GCAUUAU ((.((...((((..(((((......)))))(((....)))..))))(((((((((((((...))...((..(((((....)))))..))...)))))))))))))---.)).... ( -32.40) >DroEre_CAF1 14534 110 + 1 UGGCAAAAGUGGAGAGCAGUGGAACCUGUUGUUGAAAGACCAUGACCCCUGUUGGAACCGACGGAGCCUUGGAGCCAAUGGGCUCUGAGAAGUUUUAAAA--UUU---GAAUAAU .(((....((((..(((((......))))).((....)))))).....((((((....)))))).)))(..(((((....)))))..)............--...---....... ( -29.70) >DroMoj_CAF1 15700 111 + 1 UUGCGAAACUAGCGCCCAAUGGAACCACUUGUUGGAAUGUUUGCACGCCCGCGGGCACAGCGGGAGCUUUGGAGCCAAUGGGCUCUAAGAAAAUUAAAGC--GUUUAAACAU--U ..(((.......)))((((..(......)..))))((((((((.((((((((.......))))....(((((((((....))))))))).........))--)).)))))))--) ( -38.00) >consensus UGGCAAAAGUGGAGAGCAGUGGAACCUGUUGUUGAAAGACCAUCACGCCUGUUGGAACCGACGGAGCCUUGGAACCAAUAGGCUCUGAGAAAUUUUAAUA__UUG___GCAUUAU ........((((....(((..(......)..))).....))))...((((((((....)))))).))(((((((((....))))))))).......................... (-19.26 = -18.97 + -0.30)

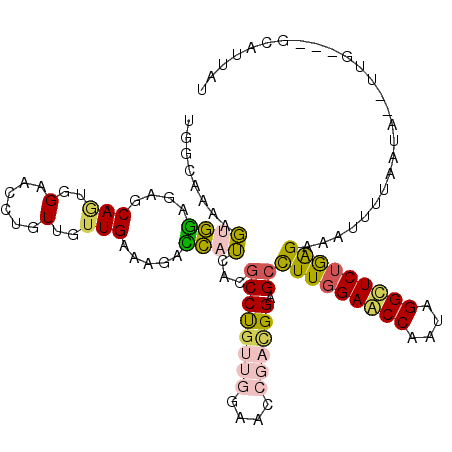
| Location | 2,863,591 – 2,863,693 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 68.48 |
| Mean single sequence MFE | -23.75 |
| Consensus MFE | -16.38 |
| Energy contribution | -16.77 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.898927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2863591 102 + 20766785 CAUCACGCCUGUUGGAACCGACGGAGCCUUGGAACCAAUAGGUUCUGGGAAAUUUUAAUAAGUUG---GCAUUCUGAUUAAGUUGUGUUCCAAAAAUCAUACCAC ...........(((((((((((.....((..(((((....)))))..)).....(((((.((...---.....)).))))))))).)))))))............ ( -26.70) >DroVir_CAF1 15091 82 + 1 CUGCACGCCCGUGGGCACAGCCGGCGCCUUUGAACCAAUAGGCUCUAAGAAAAU-AAAGC--UU--AUAAAU--UUAACAAUU-A---------------AACCA ..((..(((....)))......((.((((.(......).)))).))........-...))--..--......--.........-.---------------..... ( -13.80) >DroSec_CAF1 15224 102 + 1 CAUCACGCCUGUUGGAACCGACGGAGCCUUGGAACCAAUAGGUUCUGGGAAAUUUUAAUAGGUUG---GCUUUAUAAAUAAGUUGUGUUCCAAACGUCAUACCAC ....(((....((((((((((((((((((..(((((....)))))..)..(((((....))))))---)))))........)))).))))))).)))........ ( -27.50) >DroSim_CAF1 22095 102 + 1 CAUCACGCCUGUUGGAACCGAUGGAGCCUUGGAACCAAUAGGUUCUGGGAAAUUUUAAUAGGUUG---GCAUUAUAAAUAAGUUGUGUUCCAAACGUCAUACCAC ......((((((((....)))))).))((..(((((....)))))..))...........(((((---((.(((....)))(((........))))))).))).. ( -25.20) >DroEre_CAF1 14574 100 + 1 CAUGACCCCUGUUGGAACCGACGGAGCCUUGGAGCCAAUGGGCUCUGAGAAGUUUUAAAA--UUU---GAAUAAUAAAUAAGUUGUGUUACAAACUUCGUACUAC ((..((..((((((....))))))...((..(((((....)))))..))...........--...---.............))..)).................. ( -24.90) >DroMoj_CAF1 15740 85 + 1 UUGCACGCCCGCGGGCACAGCGGGAGCUUUGGAGCCAAUGGGCUCUAAGAAAAUUAAAGC--GUUUAAACAU--UAAAUAAUU-G---------------ACCAC ..((...(((((.......)))))...(((((((((....))))))))).........))--..........--.........-.---------------..... ( -24.40) >consensus CAUCACGCCUGUUGGAACCGACGGAGCCUUGGAACCAAUAGGCUCUGAGAAAUUUUAAUA__UUG___GCAUUAUAAAUAAGUUGUGUUCCAAAC_UCAUACCAC ......((((((((....)))))).))(((((((((....)))))))))........................................................ (-16.38 = -16.77 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:25 2006