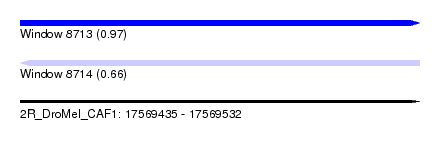
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,569,435 – 17,569,532 |
| Length | 97 |
| Max. P | 0.972481 |

| Location | 17,569,435 – 17,569,532 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 77.72 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -20.22 |
| Energy contribution | -21.20 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17569435 97 + 20766785 AGCCUGAAAAAUCCGGCCCUGGCCUUUUUUU-----UUUAUUGUUGCCUCUUGGUCAACAGCAAAGCGCUUGAGGCCGUGUAAUAACUUUCGGGCAAAUAUU .((((((((......((..(((((((....(-----(((.((((((((....)).)))))).)))).....))))))).))......))))))))....... ( -31.90) >DroSec_CAF1 75912 102 + 1 AGCCUGGAAAAUCCGGCCCUGGCCUUUUUUUUUUUUUUUUUUGUUGCCUCUUGGUCAACAGCAAAGCGCUUGAGGCCGUGUAAUAACUUUCGGGCAAAUAUU .((((((((.....(((((.(((((((.............((((((((....)).)))))).)))).))).).))))((......))))))))))....... ( -30.54) >DroSim_CAF1 65444 97 + 1 AGCCUGGAAAAUCCGGCCCUGGCCUUUUUUU-----UCUUUUGUUGCCACUUGGUCAACAGCAAAGCGCUUGAGGCCGUGUAAUAACUUUCGGGCAAAUAUU .((((((((.....(((((.(((((((....-----....((((((((....)).)))))).)))).))).).))))((......))))))))))....... ( -30.80) >DroEre_CAF1 86790 89 + 1 AGCCUGGAAAAUCCGGCCCUGGCCUUU-------------UUGUUGCCUCUUGGUCAACAGCAAAGCGCUUGAGGCCGUGUAAUAAGUUUCGGGCAAAUAUU .((((((((...(((((((.(((((((-------------((((((((....)).)))))).)))).))).).))))).).......))))))))....... ( -36.30) >DroYak_CAF1 79566 85 + 1 AGCCUGAAAAAUCCGGCCCUGGCCUUU-------------UUGUUGCCUCUUGGCCAACAGCAAAGCGCUUGAGGCCGUGUAAUAAGUU----GCAAAUAUU .............((((((.(((((((-------------((((((((....)).)))))).)))).))).).)))))(((((....))----)))...... ( -27.30) >DroPer_CAF1 95113 93 + 1 ---AU-GGAAGUCCGCCCCUGUUUUUC-AGUUCUCUGGUUCUGUUGUUCCUUAGUCAAA---CAAUUACUUAAAG-CAUGUAAUAACUUUAGCUUAAAUAUU ---..-((....))((....(((.(((-((((....(((...((((((.........))---)))).)))...))-).)).)).)))....))......... ( -8.90) >consensus AGCCUGGAAAAUCCGGCCCUGGCCUUU_UUU_____U_UUUUGUUGCCUCUUGGUCAACAGCAAAGCGCUUGAGGCCGUGUAAUAACUUUCGGGCAAAUAUU .((((((((......((..(((((((...............(((((((....)).)))))((.....))..))))))).))......))))))))....... (-20.22 = -21.20 + 0.98)

| Location | 17,569,435 – 17,569,532 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 77.72 |
| Mean single sequence MFE | -24.35 |
| Consensus MFE | -14.61 |
| Energy contribution | -16.33 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17569435 97 - 20766785 AAUAUUUGCCCGAAAGUUAUUACACGGCCUCAAGCGCUUUGCUGUUGACCAAGAGGCAACAAUAAA-----AAAAAAAGGCCAGGGCCGGAUUUUUCAGGCU .......(((.((((((....)).((((((...((.((((..(((((.((....))))))).....-----....))))))..))))))....)))).))). ( -27.20) >DroSec_CAF1 75912 102 - 1 AAUAUUUGCCCGAAAGUUAUUACACGGCCUCAAGCGCUUUGCUGUUGACCAAGAGGCAACAAAAAAAAAAAAAAAAAAGGCCAGGGCCGGAUUUUCCAGGCU .......(((.(((((((....(..(((((..(((.....)))((((.((....)))))).................)))))..)....)))))))..))). ( -26.40) >DroSim_CAF1 65444 97 - 1 AAUAUUUGCCCGAAAGUUAUUACACGGCCUCAAGCGCUUUGCUGUUGACCAAGUGGCAACAAAAGA-----AAAAAAAGGCCAGGGCCGGAUUUUCCAGGCU .......(((.(((((((....(..(((((......((((..(((((.((....))))))))))).-----......)))))..)....)))))))..))). ( -27.52) >DroEre_CAF1 86790 89 - 1 AAUAUUUGCCCGAAACUUAUUACACGGCCUCAAGCGCUUUGCUGUUGACCAAGAGGCAACAA-------------AAAGGCCAGGGCCGGAUUUUCCAGGCU .......(((...............(((((...((.((((..(((((.((....))))))).-------------))))))..)))))((.....)).))). ( -28.00) >DroYak_CAF1 79566 85 - 1 AAUAUUUGC----AACUUAUUACACGGCCUCAAGCGCUUUGCUGUUGGCCAAGAGGCAACAA-------------AAAGGCCAGGGCCGGAUUUUUCAGGCU .......((----...........((((((...((.((((..(((((.((....))))))).-------------))))))..))))))((....))..)). ( -25.40) >DroPer_CAF1 95113 93 - 1 AAUAUUUAAGCUAAAGUUAUUACAUG-CUUUAAGUAAUUG---UUUGACUAAGGAACAACAGAACCAGAGAACU-GAAAAACAGGGGCGGACUUCC-AU--- ...........((((((........)-))))).((..(((---(((......(......).....(((....))-)..))))))..))((....))-..--- ( -11.60) >consensus AAUAUUUGCCCGAAAGUUAUUACACGGCCUCAAGCGCUUUGCUGUUGACCAAGAGGCAACAAAA_A_____AAA_AAAGGCCAGGGCCGGAUUUUCCAGGCU .......(((.(((((((....(..(((((..(((.....)))((((.((....)))))).................)))))..)....)))))))..))). (-14.61 = -16.33 + 1.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:27 2006