| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,566,028 – 17,566,128 |
| Length | 100 |
| Max. P | 0.928754 |

| Location | 17,566,028 – 17,566,128 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 77.26 |
| Mean single sequence MFE | -18.13 |
| Consensus MFE | -10.65 |
| Energy contribution | -10.43 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
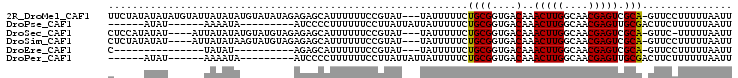
>2R_DroMel_CAF1 17566028 100 - 20766785 UUCUAUAUAUAUGUAUUAUAUAUGUAUAUAGAGAGCAUUUUUUCCGUAU---UAUUUUUCUGCGGUGACAAACUUGGCAACGAGUCGCA-GUUCCUUUUUAAUU (((((((((((((((...)))))))))))))))................---.......(((((....)..(((((....))))).)))-)............. ( -26.20) >DroPse_CAF1 86508 83 - 1 ------AUAU------AAAAUA---------AUCCCCUUUUUUCCUUAUUAUUAUUUUUCUGCGGUGACAAACUUGGCAACGAGUUGCGACUUCUUUUUUAAUU ------....------......---------.............................((((....).((((((....)))))))))............... ( -11.80) >DroSec_CAF1 72520 95 - 1 CUCCAUAUAU----AUUAUAUAUGUAUGUAGAGAGCAUUUUUUCCGUAU---UAUUUUUCUGCGGUGACAAACUUGGCAACGAGUCGCA-GUUC-UUUUUAAUU ...(((((((----((...))))))))).(((((((.......(((((.---........)))))((....(((((....)))))..))-))))-)))...... ( -21.80) >DroSim_CAF1 62048 96 - 1 CUCUAUAUAU----AUUAUAUAAGUAUGUAGAGAGCAUUUUUUCCGUAU---UAUUUUUCUGCGGUGACAAACUUGGCAACGAGUCGCA-GUUCCUUUUUAAUU ((((((((((----.........))))))))))................---.......(((((....)..(((((....))))).)))-)............. ( -23.00) >DroEre_CAF1 83256 75 - 1 C---------------UAUAU----------AGAGCAUUUUUUCCGUAU---UAUUUUUCUGCGGUGACAAACUUGGCAACGAGUCGCA-GUUCCUUUUUAAUU .---------------.(((.----------.(((......)))..)))---.......(((((....)..(((((....))))).)))-)............. ( -14.20) >DroPer_CAF1 91560 83 - 1 ------AUAU------AAAAUA---------AUCCCCUUUUUUCCUUAUUAUUAUUUUUCUGCGGUGACAAACUUGGCAACGAGUUGCGACUUCUUUUUUAAUU ------....------......---------.............................((((....).((((((....)))))))))............... ( -11.80) >consensus C_____AUAU______UAUAUA_________AGAGCAUUUUUUCCGUAU___UAUUUUUCUGCGGUGACAAACUUGGCAACGAGUCGCA_GUUCCUUUUUAAUU ............................................................((((....)..(((((....))))).)))............... (-10.65 = -10.43 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:24 2006