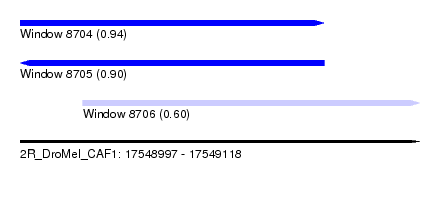
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,548,997 – 17,549,118 |
| Length | 121 |
| Max. P | 0.938585 |

| Location | 17,548,997 – 17,549,089 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.22 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -21.01 |
| Energy contribution | -21.98 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17548997 92 + 20766785 CU--------------CCG--GGCCACCGGCUUCUCCCGUUGGCCACAUUUAAAUAUUUGCGCACGGAAUUAUUUUCCCUGGCAUAAAGUGUGGCCAGGUAGACGUUU ..--------------(((--(....))))..(((.((..(((((((((((.......(((.((.((((.....)))).)))))..))))))))))))).)))..... ( -33.80) >DroSec_CAF1 55477 92 + 1 CU--------------CUG--GGCCACCGGCUUCUCCCGUUGGCCACAUUUAAAUAUUUGCGCACGGAAUUAUUUUCCCUGGCAUAAAGUGUGGCCAGGUAGACGUUU ..--------------(((--(....))))..(((.((..(((((((((((.......(((.((.((((.....)))).)))))..))))))))))))).)))..... ( -31.60) >DroEre_CAF1 63487 92 + 1 CU--------------CUG--CGCCACCGGCUUCUCCCGUUGGCCACAUUUAAAUAUUUGCGCACGGAAUUAUUUUCCCUGGCAUAAAGUGUGGCCAGGUAGACGUUU .(--------------(((--((((...)))........((((((((((((.......(((.((.((((.....)))).)))))..)))))))))))))))))..... ( -32.70) >DroYak_CAF1 58549 92 + 1 CU--------------CUG--GGCCACCGGCUUCUCCCGUUGGCCACAUUUAAAUAUUUGCGCACGGAAUUAUUUUCCCUGGCAUAAAGUGUGGCCAGGUAGACGUUU ..--------------(((--(....))))..(((.((..(((((((((((.......(((.((.((((.....)))).)))))..))))))))))))).)))..... ( -31.60) >DroAna_CAF1 57893 103 + 1 CUUCUGCCUCCUCCGCCAG--GACCACCCACGUC---CCUGGGCCACAUUUAAAUAUUUGCGCUUGGAAUUAUUUUCGCCGGCAUAAAGUGUGGCCAGGUAGACAUUU ..(((((((.......(((--(..(......)..---))))((((((((((.......(((...(((((....)))))...)))..)))))))))))))))))..... ( -32.90) >DroPer_CAF1 72790 85 + 1 ---------------GGAGGGUGGCACGCG-------CCUCAGCCGCAUUUAAAUAUUUGCGCAUGGAAUUAUUUUCUCUG-CGAAAAUGGCGGCCAGGCAGACGUUU ---------------..........(((((-------(((..(((((.......(((((.((((.((((.....)))).))-)).)))))))))).)))).).))).. ( -25.81) >consensus CU______________CUG__GGCCACCGGCUUCUCCCGUUGGCCACAUUUAAAUAUUUGCGCACGGAAUUAUUUUCCCUGGCAUAAAGUGUGGCCAGGUAGACGUUU ......................(((..(((......)))..((((((((((.......(((.((.((((.....)))).)))))..)))))))))).)))........ (-21.01 = -21.98 + 0.98)

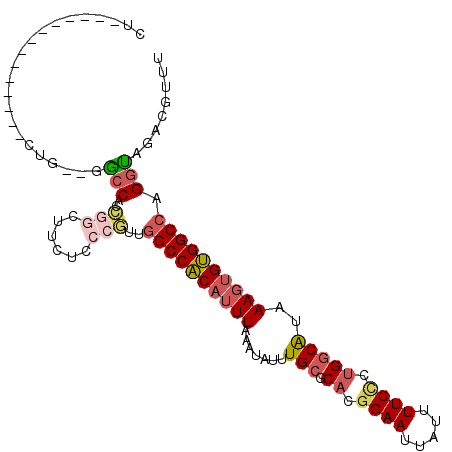
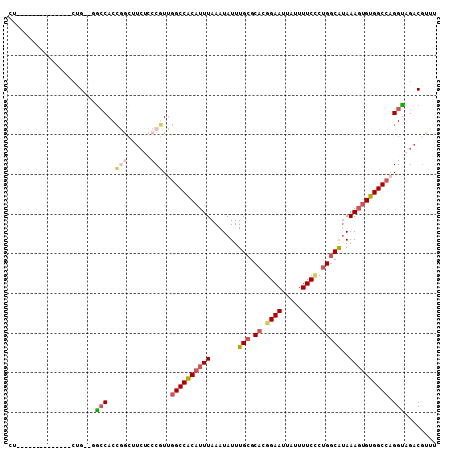
| Location | 17,548,997 – 17,549,089 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.22 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -19.53 |
| Energy contribution | -21.20 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17548997 92 - 20766785 AAACGUCUACCUGGCCACACUUUAUGCCAGGGAAAAUAAUUCCGUGCGCAAAUAUUUAAAUGUGGCCAACGGGAGAAGCCGGUGGCC--CGG--------------AG .....(((.((((((((((.(((((((((.((((.....)))).)).)))......))))))))))))..)).)))..((((....)--)))--------------.. ( -32.50) >DroSec_CAF1 55477 92 - 1 AAACGUCUACCUGGCCACACUUUAUGCCAGGGAAAAUAAUUCCGUGCGCAAAUAUUUAAAUGUGGCCAACGGGAGAAGCCGGUGGCC--CAG--------------AG ....(.(((((((((((((.(((((((((.((((.....)))).)).)))......))))))))))))..((......)))))))).--...--------------.. ( -30.80) >DroEre_CAF1 63487 92 - 1 AAACGUCUACCUGGCCACACUUUAUGCCAGGGAAAAUAAUUCCGUGCGCAAAUAUUUAAAUGUGGCCAACGGGAGAAGCCGGUGGCG--CAG--------------AG ...((.(((((((((((((.(((((((((.((((.....)))).)).)))......))))))))))))..((......)))))))))--...--------------.. ( -33.00) >DroYak_CAF1 58549 92 - 1 AAACGUCUACCUGGCCACACUUUAUGCCAGGGAAAAUAAUUCCGUGCGCAAAUAUUUAAAUGUGGCCAACGGGAGAAGCCGGUGGCC--CAG--------------AG ....(.(((((((((((((.(((((((((.((((.....)))).)).)))......))))))))))))..((......)))))))).--...--------------.. ( -30.80) >DroAna_CAF1 57893 103 - 1 AAAUGUCUACCUGGCCACACUUUAUGCCGGCGAAAAUAAUUCCAAGCGCAAAUAUUUAAAUGUGGCCCAGG---GACGUGGGUGGUC--CUGGCGGAGGAGGCAGAAG ...(((((.((((((((((.((((.....(((..............))).......)))))))))))((((---..((....))..)--)))....)))))))).... ( -30.84) >DroPer_CAF1 72790 85 - 1 AAACGUCUGCCUGGCCGCCAUUUUCG-CAGAGAAAAUAAUUCCAUGCGCAAAUAUUUAAAUGCGGCUGAGG-------CGCGUGCCACCCUCC--------------- ..((((..((((((((((.((((.((-((..(((.....)))..)))).........)))))))))).)))-------)))))..........--------------- ( -25.80) >consensus AAACGUCUACCUGGCCACACUUUAUGCCAGGGAAAAUAAUUCCGUGCGCAAAUAUUUAAAUGUGGCCAACGGGAGAAGCCGGUGGCC__CAG______________AG ......(((((((((((((.(((((((((.((((.....)))).)).)))......))))))))))))..((......)))))))....................... (-19.53 = -21.20 + 1.67)

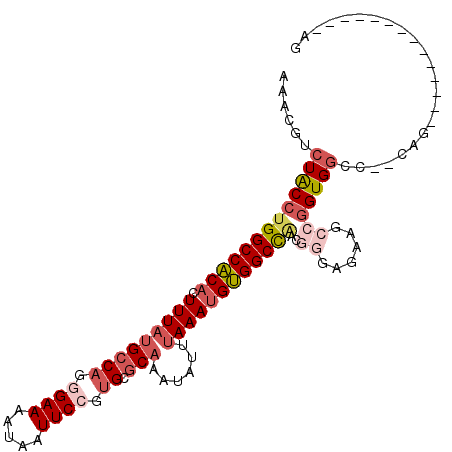
| Location | 17,549,016 – 17,549,118 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 84.30 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -20.23 |
| Energy contribution | -21.07 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
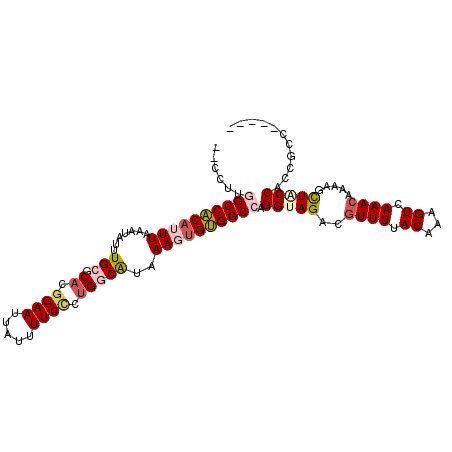
>2R_DroMel_CAF1 17549016 102 + 20766785 CCCGUUGGCCACAUUUAAAUAUUUGCGCACGGAAUUAUUUUCCCUGGCAUAAAGUGUGGCCAGGUAGACGUUUUACAAAGUCAAACAAAAGCUACCACCUCC----- .....(((((((((((.......(((.((.((((.....)))).)))))..)))))))))))(((((..((((.((...)).)))).....)))))......----- ( -31.40) >DroPse_CAF1 67858 104 + 1 --CCUCAGCCGCAUUUAAAUAUUUGCGCAUGGAAUUAUUUUCUCUG-CGAAAAUUGCGGCCAGGCAGACGUUUUACAAAGUCAAACAAAAGUUACCAACGUUAUUUG --(((..((((((.......((((.((((.((((.....)))).))-)).)))))))))).)))..((((((..((..............))....))))))..... ( -23.35) >DroEre_CAF1 63506 102 + 1 CCCGUUGGCCACAUUUAAAUAUUUGCGCACGGAAUUAUUUUCCCUGGCAUAAAGUGUGGCCAGGUAGACGUUUUACAAAGUCAAACAAAAGCUGCCACCGCC----- .....(((((((((((.......(((.((.((((.....)))).)))))..)))))))))))(((((..((((.((...)).)))).....)))))......----- ( -31.40) >DroYak_CAF1 58568 102 + 1 CCCGUUGGCCACAUUUAAAUAUUUGCGCACGGAAUUAUUUUCCCUGGCAUAAAGUGUGGCCAGGUAGACGUUUUACAAAGUCAAACAAAAGCUGCCACCGCC----- .....(((((((((((.......(((.((.((((.....)))).)))))..)))))))))))(((((..((((.((...)).)))).....)))))......----- ( -31.40) >DroAna_CAF1 57925 104 + 1 --CCUGGGCCACAUUUAAAUAUUUGCGCUUGGAAUUAUUUUCGCCGGCAUAAAGUGUGGCCAGGUAGACAUUUAACAAAGUCAAACAGAAGUUGCCACCGCUUGUU- --(((((.((((((((.......(((...(((((....)))))...)))..)))))))))))))..((((.....(((..((.....))..)))........))))- ( -23.52) >DroPer_CAF1 72805 104 + 1 --CCUCAGCCGCAUUUAAAUAUUUGCGCAUGGAAUUAUUUUCUCUG-CGAAAAUGGCGGCCAGGCAGACGUUUUACAAAGUCAAACAAAAGUUACCAACGUUAUUUG --(((..(((((.......(((((.((((.((((.....)))).))-)).)))))))))).)))..((((((..((..............))....))))))..... ( -22.75) >consensus __CCUUGGCCACAUUUAAAUAUUUGCGCACGGAAUUAUUUUCCCUGGCAUAAAGUGUGGCCAGGUAGACGUUUUACAAAGUCAAACAAAAGCUACCACCGCC_____ ......((((((((((.......(((.((.((((.....)))).)))))..)))))))))).(((((..((((.((...)).)))).....)))))........... (-20.23 = -21.07 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:19 2006