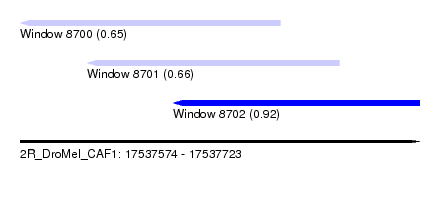
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,537,574 – 17,537,723 |
| Length | 149 |
| Max. P | 0.916472 |

| Location | 17,537,574 – 17,537,671 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 76.39 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -14.92 |
| Energy contribution | -14.70 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17537574 97 - 20766785 AACGUAUGCGUGAUAUUGAAACAAUGUGAAAAUUGCGCAUACGCACAGUUGUACCACC--UCGCACGCAUUGUUCCCUGGGCAAGC---AA-ACCAAGUCUUU ..((((((((..((.((.(.......).)).))..))))))))...............--......((.(((((.....)))))))---..-........... ( -20.60) >DroPse_CAF1 55996 73 - 1 UACGUAUGCGUAAUAUUGAAACAAUGUGAAAAUUGCGCAUACGCACAGUUGUUCCGCU--UCACACGCAAGACUC---------------------------- ..((((((((((((.((.(.......).)).)))))))))))).......(((..((.--......))..)))..---------------------------- ( -17.00) >DroEre_CAF1 51719 97 - 1 AACGUAUGCGUGAUAUUGAAACAAUGUGAAAAUUGCGCAUACGCACAGUUGUGCCACC--UCGCACGCAUUGUUCCCUGGGCAAGC---AA-ACCAAGUCCUU ..((((((((..((.((.(.......).)).))..)))))))).((((((((((....--..))))).))))).....((((....---..-.....)))).. ( -26.50) >DroYak_CAF1 46670 99 - 1 AACGUAUGCGUGAUAUUGAAACAAUGUGAAAAUUGCGCAUACGCACAGUUGUACCACCACUCGCACGCAUUGUUCCCUGGGCAAGC---AA-ACCAAGUACUU ...(((((((..((.((.(.......).)).))..)))))))((..(((.(.....).))).))..((.(((((.....)))))))---..-........... ( -21.20) >DroAna_CAF1 47308 101 - 1 AACGUAUGCGUGAUAUUGAAACAAUGUGAAAAUUGCGCAUACGCACAGUUGUACCACU--UCGCACGCAAUGUUUUCUGGGUGAGUUCCACCCCCAGGUCCCC (((((.((((((((((((...))))))...(((((.((....)).)))))........--...))))))))))).((((((.(........)))))))..... ( -26.10) >DroPer_CAF1 60857 73 - 1 UACGUAUGCGUAAUAUUGAAACAAUGUGAAAAUUGCGCAUACGCACAGUUGUUCCGCU--UCACACGCAAGACUC---------------------------- ..((((((((((((.((.(.......).)).)))))))))))).......(((..((.--......))..)))..---------------------------- ( -17.00) >consensus AACGUAUGCGUGAUAUUGAAACAAUGUGAAAAUUGCGCAUACGCACAGUUGUACCACC__UCGCACGCAAUGUUCCCUGGGCAAGC___AA_ACCAAGUCCUU ..((((((((((((.((.(.......).)).))))))))))))............................................................ (-14.92 = -14.70 + -0.22)

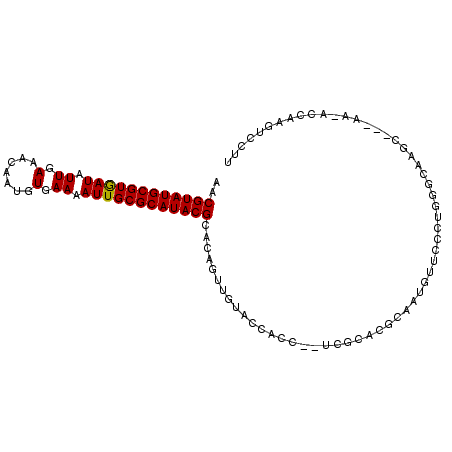
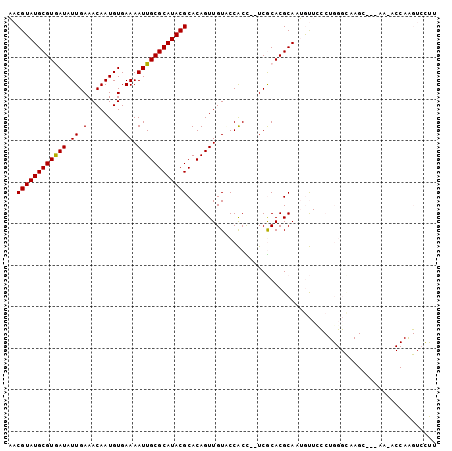
| Location | 17,537,599 – 17,537,693 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 90.56 |
| Mean single sequence MFE | -20.60 |
| Consensus MFE | -17.79 |
| Energy contribution | -17.98 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17537599 94 - 20766785 CCAACAUGCACAGUCACAAUUUAACGUAUGCGUGAUAUUGAAACAAUGUGAAAAUUGCGCAUACGCACAGUUGUACCACC--UCGCACGCAUUGUU ..(((((((...((.((((((...((((((((..((.((.(.......).)).))..))))))))...))))))))(...--..)...))).)))) ( -20.90) >DroSec_CAF1 44018 94 - 1 CCCACAUGCACAGUCACAAUUUAACGUAUGCGUGAUAUUGAAACAAUGUGAAAAUUGCGCAUACGCACAGUUGUAUCACC--UCGCACACAUUGUU ....((((((..((.........))...)))))).......(((((((((..(((((.((....)).)))))((......--..)).))))))))) ( -20.60) >DroSim_CAF1 45450 94 - 1 CCCACAUGCACAGUCACAAUUUAACGUAUGCGUGAUAUUGAAACAAUGUGAAAAUUGCGCAUACGCACAGUUGUACCACC--UCGCACGCAUUGUU ......(((...((.((((((...((((((((..((.((.(.......).)).))..))))))))...))))))...)).--..)))......... ( -20.30) >DroEre_CAF1 51744 94 - 1 CCCACAUACACAGUCACAAUUUAACGUAUGCGUGAUAUUGAAACAAUGUGAAAAUUGCGCAUACGCACAGUUGUGCCACC--UCGCACGCAUUGUU ........................((((((((..((.((.(.......).)).))..)))))))).((((((((((....--..))))).))))). ( -23.10) >DroYak_CAF1 46695 96 - 1 UGAACAUACACAGUUACAAUUUAACGUAUGCGUGAUAUUGAAACAAUGUGAAAAUUGCGCAUACGCACAGUUGUACCACCACUCGCACGCAUUGUU ............(.(((((((...((((((((..((.((.(.......).)).))..))))))))...))))))).)................... ( -20.50) >DroAna_CAF1 47337 83 - 1 ---------UCAC--UCACUUUAACGUAUGCGUGAUAUUGAAACAAUGUGAAAAUUGCGCAUACGCACAGUUGUACCACU--UCGCACGCAAUGUU ---------....--.......(((((.((((((((((((...))))))...(((((.((....)).)))))........--...))))))))))) ( -18.20) >consensus CCCACAUGCACAGUCACAAUUUAACGUAUGCGUGAUAUUGAAACAAUGUGAAAAUUGCGCAUACGCACAGUUGUACCACC__UCGCACGCAUUGUU ................((((...(((....)))...)))).(((((((((..(((((.((....)).)))))((..........)).))))))))) (-17.79 = -17.98 + 0.19)

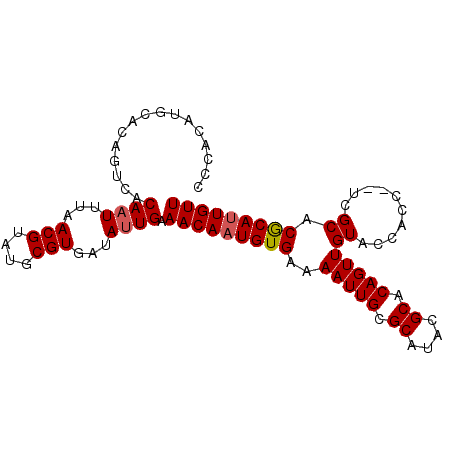

| Location | 17,537,631 – 17,537,723 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 70.40 |
| Mean single sequence MFE | -10.05 |
| Consensus MFE | -7.82 |
| Energy contribution | -7.60 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17537631 92 - 20766785 UCCCCAAUCCAA----CUCC--UCCCCUCCCACCCACCAACAUGCACAGUCACAAUUUAACGUAUGCGUGAUAUUGAAACAAUGUGAAAAUUGCGCAU ............----....--...................((((.......((((...(((....)))...))))...((((......)))).)))) ( -10.10) >DroPse_CAF1 56029 80 - 1 ACCCCAACCCAACCUUAAGCACCCCGUU-----UCACCCCC-------------CUCCUACGUAUGCGUAAUAUUGAAACAAUGUGAAAAUUGCGCAU ..................(((.....((-----((((....-------------....((((....))))..((((...))))))))))..))).... ( -9.80) >DroSec_CAF1 44050 88 - 1 UCCCCAAUCCACA---CACC--CCCCCU-----CCACCCACAUGCACAGUCACAAUUUAACGUAUGCGUGAUAUUGAAACAAUGUGAAAAUUGCGCAU .............---....--......-----........((((.......((((...(((....)))...))))...((((......)))).)))) ( -10.10) >DroSim_CAF1 45482 90 - 1 UCCCCAAUCCACC---AACCCCUCCCCU-----CCACCCACAUGCACAGUCACAAUUUAACGUAUGCGUGAUAUUGAAACAAUGUGAAAAUUGCGCAU .............---............-----........((((.......((((...(((....)))...))))...((((......)))).)))) ( -10.10) >DroYak_CAF1 46729 82 - 1 UCCCCAAU----------------AAACCUUCCUCCUGAACAUACACAGUUACAAUUUAACGUAUGCGUGAUAUUGAAACAAUGUGAAAAUUGCGCAU ........----------------....................(((((((.((((...(((....)))...)))).)))..))))............ ( -10.40) >DroPer_CAF1 60890 80 - 1 ACCCCAAUACAACCUUAAGCACCCCGUU-----UCACCCCC-------------CUCCUACGUAUGCGUAAUAUUGAAACAAUGUGAAAAUUGCGCAU ..................(((.....((-----((((....-------------....((((....))))..((((...))))))))))..))).... ( -9.80) >consensus UCCCCAAUCCAAC___AACC__CCCCCU_____CCACCCACAUGCACAGUCACAAUUUAACGUAUGCGUGAUAUUGAAACAAUGUGAAAAUUGCGCAU ...............................................................(((((((((.((.(.......).)).))))))))) ( -7.82 = -7.60 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:16 2006