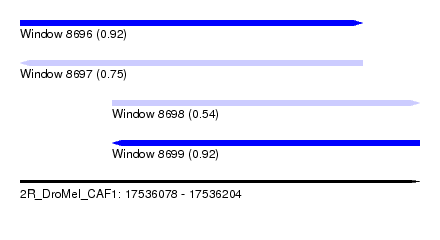
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,536,078 – 17,536,204 |
| Length | 126 |
| Max. P | 0.919841 |

| Location | 17,536,078 – 17,536,186 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 81.61 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -16.85 |
| Energy contribution | -16.38 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
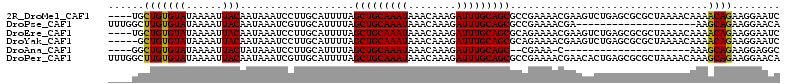
>2R_DroMel_CAF1 17536078 108 + 20766785 ----UGCUGUGUAUAAAAUUACAAUAAAUCCUUGCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGCGCCGAAAACGAAGUCUGAGCGCGCUAAAACAAAACAGAAGGAAUC ----.....((((......)))).....(((((...((((((((((((((........)))))))(((((((........)).).)))))))))))(......))))))... ( -23.70) >DroPse_CAF1 54509 92 + 1 UUUGGCUUGUGUAUAAAAUUACAAUAAAUCGUUGCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGCGCCGAAAACGA--------------------AAGCAGAAGGAACA ....((((.((((......)))).....(((((........(((((((((........)))))))))......)))))--------------------)))).......... ( -22.94) >DroEre_CAF1 50243 108 + 1 ----UGCUGUGUAUAAAAUUACAAUAAAUCCUUGCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGCGCAGAAAACGAAGUCUGAGCGCGCUAAAACAAAACAGAAGGAAUC ----.....((((......)))).....(((((...((((((((((((((........)))))))(((((((........)))..)))))))))))(......))))))... ( -24.40) >DroYak_CAF1 45195 107 + 1 -----GCUGUGUAUAAAAUUACAAUAAAUCCUUGCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGCGCAGAAAACGAAGUCUGAGCGCGCUAAAACAAAACAGAAGGAAUC -----....((((......)))).....(((((...((((((((((((((........)))))))(((((((........)))..)))))))))))(......))))))... ( -24.40) >DroAna_CAF1 46119 84 + 1 ----GGCUGUGUAUAAAAUUACUAUAAAUCCUUGCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGC--CGAAA-C---------------------AAAGCAGAAGGAGGC ----.(((..(((......)))......(((((((.(((..(((((((((........)))))))))--..)))-.---------------------...)))..))))))) ( -21.60) >DroPer_CAF1 59320 112 + 1 UUUGGCUUGUGUAUAAAAUUACAAUAAAUCGUUGCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGCGCCGAAAACGAACACUGAGCGCGCUAAAACAAAGCAGAAGGAACA ((((((.((((((......)))......(((((........(((((((((........)))))))))......))))).......))).))))))................. ( -25.34) >consensus ____GGCUGUGUAUAAAAUUACAAUAAAUCCUUGCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGCGCCGAAAACGAAGUCUGAGCGCGCUAAAACAAAACAGAAGGAAUC ......(((((((......)))...................(((((((((........))))))))).................................))))........ (-16.85 = -16.38 + -0.47)
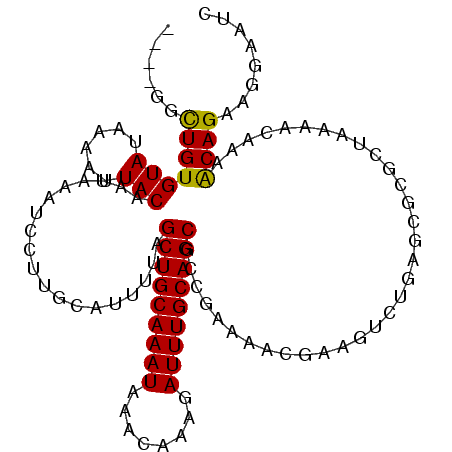
| Location | 17,536,078 – 17,536,186 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 81.61 |
| Mean single sequence MFE | -24.02 |
| Consensus MFE | -14.05 |
| Energy contribution | -14.22 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17536078 108 - 20766785 GAUUCCUUCUGUUUUGUUUUAGCGCGCUCAGACUUCGUUUUCGGCGCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGCAAGGAUUUAUUGUAAUUUUAUACACAGCA---- ...((((..(((...(((((((((((((.(((......))).)))))(((((((........)))))))))))))))))))))).....((((......)))).....---- ( -26.70) >DroPse_CAF1 54509 92 - 1 UGUUCCUUCUGCUU--------------------UCGUUUUCGGCGCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGCAACGAUUUAUUGUAAUUUUAUACACAAGCCAAA ..............--------------------........((((((((((((........)))))))))((((((((((.......)))).)))))).......)))... ( -20.10) >DroEre_CAF1 50243 108 - 1 GAUUCCUUCUGUUUUGUUUUAGCGCGCUCAGACUUCGUUUUCUGCGCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGCAAGGAUUUAUUGUAAUUUUAUACACAGCA---- ...((((..(((...((((((((((((..((((...))))...))))(((((((........)))))))))))))))))))))).....((((......)))).....---- ( -25.70) >DroYak_CAF1 45195 107 - 1 GAUUCCUUCUGUUUUGUUUUAGCGCGCUCAGACUUCGUUUUCUGCGCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGCAAGGAUUUAUUGUAAUUUUAUACACAGC----- ...((((..(((...((((((((((((..((((...))))...))))(((((((........)))))))))))))))))))))).....((((......))))....----- ( -25.70) >DroAna_CAF1 46119 84 - 1 GCCUCCUUCUGCUUU---------------------G-UUUCG--GCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGCAAGGAUUUAUAGUAAUUUUAUACACAGCC---- ((.((((..(((...---------------------(-(((..--(((((((((........)))))))))..)))))))))))......(((......)))...)).---- ( -21.60) >DroPer_CAF1 59320 112 - 1 UGUUCCUUCUGCUUUGUUUUAGCGCGCUCAGUGUUCGUUUUCGGCGCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGCAACGAUUUAUUGUAAUUUUAUACACAAGCCAAA ..........((((.(((((((((((((.((........)).)))))(((((((........)))))))))))))))............((((......)))).)))).... ( -24.30) >consensus GAUUCCUUCUGCUUUGUUUUAGCGCGCUCAGACUUCGUUUUCGGCGCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGCAAGGAUUUAUUGUAAUUUUAUACACAGCC____ .............................................(((((((((........)))))))))((((((((((.......)))).))))))............. (-14.05 = -14.22 + 0.17)

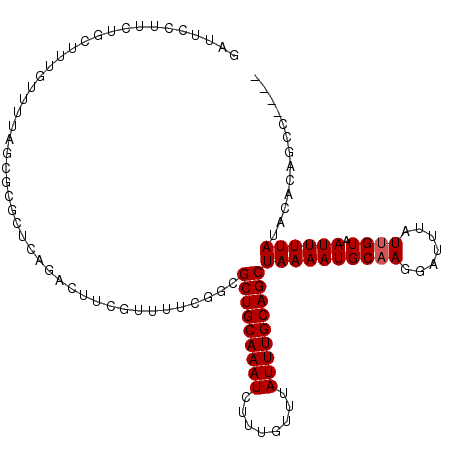
| Location | 17,536,107 – 17,536,204 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.90 |
| Mean single sequence MFE | -25.11 |
| Consensus MFE | -18.73 |
| Energy contribution | -18.92 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17536107 97 + 20766785 GCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGCGCCGAAAACGAAGUCUGAGCGCGCUAAAACAAAACAGAAGGAAUCGGAAGGCCAAAGUGUAUG---------------------- (((((((.(((((((((........)))))))))(((.....(((..((((.................)))).....)))...))).)))))))...---------------------- ( -25.53) >DroSec_CAF1 42650 97 + 1 GCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGCGCCGAAAACGAAGUCUGAGCGCGCUAAAACAAAACAGAAGGAAUCGGAAGGCCAAAGUGUAUG---------------------- (((((((.(((((((((........)))))))))(((.....(((..((((.................)))).....)))...))).)))))))...---------------------- ( -25.53) >DroSim_CAF1 43984 97 + 1 GCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGCGCCGAAAACGAAGUCUGAGCGCGCUAAAACAAAACAGAAGGAAUCGGAAGGCCAAAGUGUAUG---------------------- (((((((.(((((((((........)))))))))(((.....(((..((((.................)))).....)))...))).)))))))...---------------------- ( -25.53) >DroEre_CAF1 50272 97 + 1 GCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGCGCAGAAAACGAAGUCUGAGCGCGCUAAAACAAAACAGAAGGAAUCGGAAGGCCGAAGUAUGUG---------------------- (((((((((((((((((........)))))))(((((((........)))..)))))))))))..............((((....))))....))).---------------------- ( -24.60) >DroYak_CAF1 45223 97 + 1 GCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGCGCAGAAAACGAAGUCUGAGCGCGCUAAAACAAAACAGAAGGAAUCGGAAGGCCGAAGUAUAUG---------------------- ...((((((((((((((........)))))))(((((((........)))..)))))))))))..............((((....))))........---------------------- ( -24.00) >DroPer_CAF1 59353 116 + 1 GCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGCGCCGAAAACGAACACUGAGCGCGCUAAAACAAAGCAGAAGGAACAGGGA---UAUAGGAUAUAAACGAUUCGAGGCACUAAGCAC ((......(((((((((........)))))))))(((...........(((..(.((((.......))).)...)..)))(((---(...(........).))))..))).....)).. ( -25.50) >consensus GCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGCGCCGAAAACGAAGUCUGAGCGCGCUAAAACAAAACAGAAGGAAUCGGAAGGCCAAAGUAUAUG______________________ ...((((((((((((((........)))))))((((................)))))))))))..........((...(....).))................................ (-18.73 = -18.92 + 0.20)

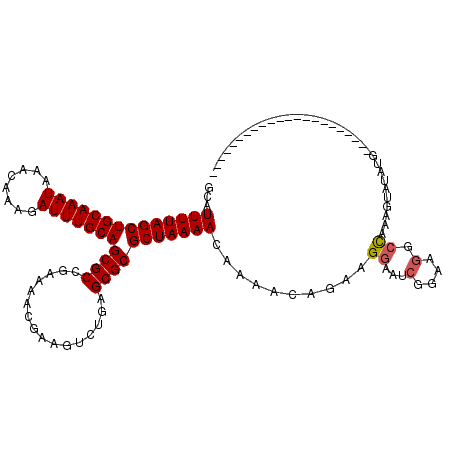

| Location | 17,536,107 – 17,536,204 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 86.90 |
| Mean single sequence MFE | -23.52 |
| Consensus MFE | -20.47 |
| Energy contribution | -21.13 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17536107 97 - 20766785 ----------------------CAUACACUUUGGCCUUCCGAUUCCUUCUGUUUUGUUUUAGCGCGCUCAGACUUCGUUUUCGGCGCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGC ----------------------........((((....)))).............(((((((((((((.(((......))).)))))(((((((........))))))))))))))).. ( -22.90) >DroSec_CAF1 42650 97 - 1 ----------------------CAUACACUUUGGCCUUCCGAUUCCUUCUGUUUUGUUUUAGCGCGCUCAGACUUCGUUUUCGGCGCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGC ----------------------........((((....)))).............(((((((((((((.(((......))).)))))(((((((........))))))))))))))).. ( -22.90) >DroSim_CAF1 43984 97 - 1 ----------------------CAUACACUUUGGCCUUCCGAUUCCUUCUGUUUUGUUUUAGCGCGCUCAGACUUCGUUUUCGGCGCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGC ----------------------........((((....)))).............(((((((((((((.(((......))).)))))(((((((........))))))))))))))).. ( -22.90) >DroEre_CAF1 50272 97 - 1 ----------------------CACAUACUUCGGCCUUCCGAUUCCUUCUGUUUUGUUUUAGCGCGCUCAGACUUCGUUUUCUGCGCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGC ----------------------........((((....)))).............((((((((((((..((((...))))...))))(((((((........))))))))))))))).. ( -24.00) >DroYak_CAF1 45223 97 - 1 ----------------------CAUAUACUUCGGCCUUCCGAUUCCUUCUGUUUUGUUUUAGCGCGCUCAGACUUCGUUUUCUGCGCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGC ----------------------........((((....)))).............((((((((((((..((((...))))...))))(((((((........))))))))))))))).. ( -24.00) >DroPer_CAF1 59353 116 - 1 GUGCUUAGUGCCUCGAAUCGUUUAUAUCCUAUA---UCCCUGUUCCUUCUGCUUUGUUUUAGCGCGCUCAGUGUUCGUUUUCGGCGCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGC (..((.(((((...((((.(..((((...))))---..)..)))).....(((.......)))))))).))..).((((((....(((((((((........))))))))).)))))). ( -24.40) >consensus ______________________CAUACACUUUGGCCUUCCGAUUCCUUCUGUUUUGUUUUAGCGCGCUCAGACUUCGUUUUCGGCGCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGC ..............................((((....)))).............((((((((((((..(((......)))..))))(((((((........))))))))))))))).. (-20.47 = -21.13 + 0.67)
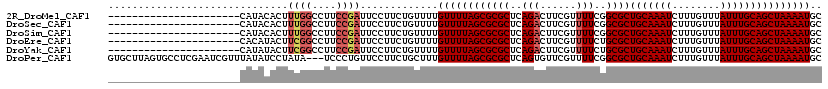
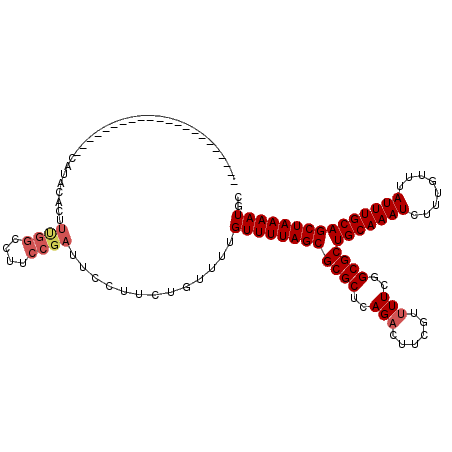
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:13 2006