
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
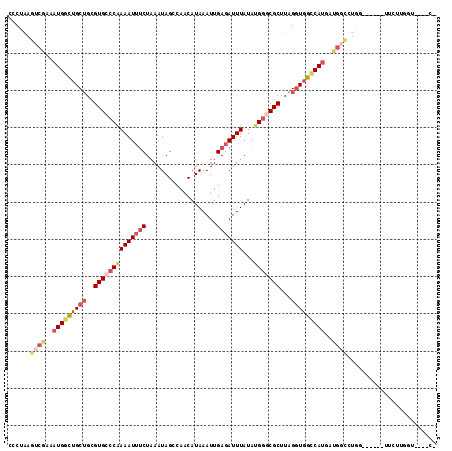
| Location | 17,519,383 – 17,519,492 |
| Length | 109 |
| Max. P | 0.886948 |

| Location | 17,519,383 – 17,519,492 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.13 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -20.29 |
| Energy contribution | -22.21 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
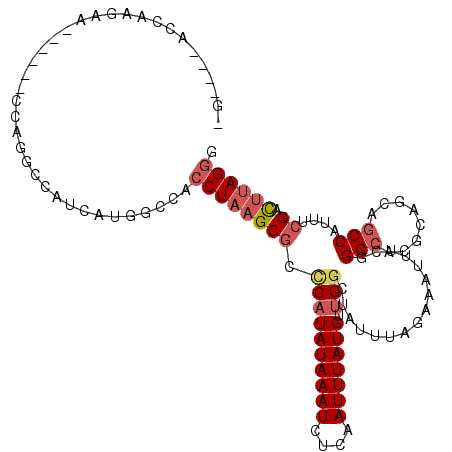
>2R_DroMel_CAF1 17519383 109 + 20766785 CCCUAAGUCGAAAUGGCUGCUGCGUGCCCGAAAUUUCUAAAUAGCCAACAUAAAUUGAGAUUUAUAUGGGCGCUUAGGUGGCCAUGAUGGCCUGG------UUCUUGGUCCUGCU ..(((.((((..(((((..((..((((((((((((((...................)))))))...)))))))...))..)))))..)))).)))------.............. ( -34.21) >DroVir_CAF1 42825 92 + 1 GCCUA-ACCCAAAUGGCAGCAGCGUGCCCAAAAUGCCUAAAUAGGCAACAUAAAUUGAGAUUUAUAUGGGCGCUUAGUUGGGCAGGUC-UU------------C---------CA (((((-((.......((....))(((((((...(((((....)))))..((((((....)))))).)))))))...))))))).((..-..------------)---------). ( -33.70) >DroPse_CAF1 35689 115 + 1 ACCUAAGCCGAAAUGGUUGCUGCGUGCCCAAAAUUUCUAAAUAGCCAACAUAAAUUGAGAUUUAUAUGGCCGCUUAGGUGGCCAUCAUGACUUGCCCAAUGAUUUUGGUCCUUUG ......((((((((.((((..(((......(((((((...................)))))))..((((((((....)))))))).......))).)))).))))))))...... ( -30.01) >DroSec_CAF1 25847 103 + 1 CCCUAAGUCGAAAUGGCUGCUGCGUGCCCGAAAUUUCUAAAUAGCCAACAUAAAUUGAGAUUUAUAUGAGCGCUUAGGUGGCCAUGAUGGCCUGG------UUCUUGGU------ ..(((.((((..(((((..((((((.....(((((((...................)))))))......))))...))..)))))..)))).)))------........------ ( -27.31) >DroEre_CAF1 33278 109 + 1 CCCUAAGUCGAAAUGGCUGCUGCGUGCCCGAAAUUUCUAAAUAGCCAACAUAAAUUGAGAUUUAUAUGGGCGCUUAGGUGGUCAUGAUGGCCUGG------UUGUAGGUCCUUCU ............(((((..((..((((((((((((((...................)))))))...)))))))...))..)))))((.((((((.------...))))))..)). ( -32.01) >DroAna_CAF1 31719 91 + 1 CCCUGAGUCGGAAUGGCUGCUGCGUGCCCAAAAUUUCUAAAUAGCCGACAUAAAUUGAGAUUUAUAAGGCCGCUUAGGUGGCCAUCCCGGG------------------------ .......((((.(((((..(((((.(((..(((((((...................)))))))....))))))...))..))))).)))).------------------------ ( -29.61) >consensus CCCUAAGUCGAAAUGGCUGCUGCGUGCCCAAAAUUUCUAAAUAGCCAACAUAAAUUGAGAUUUAUAUGGGCGCUUAGGUGGCCAUGAUGGCCUGG______UUCUUGGU____C_ ......((((..(((((((((..((((((((((((((...................)))))))...)))))))...)))))))))..))))........................ (-20.29 = -22.21 + 1.92)


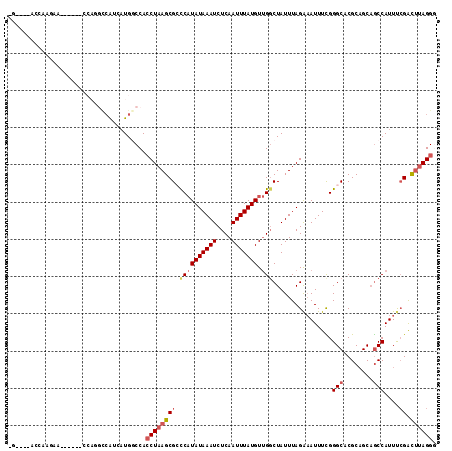
| Location | 17,519,383 – 17,519,492 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.13 |
| Mean single sequence MFE | -30.63 |
| Consensus MFE | -12.55 |
| Energy contribution | -13.47 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17519383 109 - 20766785 AGCAGGACCAAGAA------CCAGGCCAUCAUGGCCACCUAAGCGCCCAUAUAAAUCUCAAUUUAUGUUGGCUAUUUAGAAAUUUCGGGCACGCAGCAGCCAUUUCGACUUAGGG ....((........------)).((((.....)))).((((((((((.((((((((....)))))))).)))......(((((....(((........))))))))).)))))). ( -32.40) >DroVir_CAF1 42825 92 - 1 UG---------G------------AA-GACCUGCCCAACUAAGCGCCCAUAUAAAUCUCAAUUUAUGUUGCCUAUUUAGGCAUUUUGGGCACGCUGCUGCCAUUUGGGU-UAGGC ..---------.------------..-..(((((((((....(.((((((((((((....))))))))(((((....)))))....)))).)((....))...))))).-)))). ( -29.80) >DroPse_CAF1 35689 115 - 1 CAAAGGACCAAAAUCAUUGGGCAAGUCAUGAUGGCCACCUAAGCGGCCAUAUAAAUCUCAAUUUAUGUUGGCUAUUUAGAAAUUUUGGGCACGCAGCAACCAUUUCGGCUUAGGU ....((.(((...((((.((.....)))))))))))((((((((((((((((((((....))))))).))))).....(((((.....((.....))....))))).)))))))) ( -32.20) >DroSec_CAF1 25847 103 - 1 ------ACCAAGAA------CCAGGCCAUCAUGGCCACCUAAGCGCUCAUAUAAAUCUCAAUUUAUGUUGGCUAUUUAGAAAUUUCGGGCACGCAGCAGCCAUUUCGACUUAGGG ------........------...((((.....)))).((((((((((.((((((((....)))))))).)))......(((((....(((........))))))))).)))))). ( -29.90) >DroEre_CAF1 33278 109 - 1 AGAAGGACCUACAA------CCAGGCCAUCAUGACCACCUAAGCGCCCAUAUAAAUCUCAAUUUAUGUUGGCUAUUUAGAAAUUUCGGGCACGCAGCAGCCAUUUCGACUUAGGG .((.((.(((....------..))))).)).......((((((((((.((((((((....)))))))).)))......(((((....(((........))))))))).)))))). ( -30.00) >DroAna_CAF1 31719 91 - 1 ------------------------CCCGGGAUGGCCACCUAAGCGGCCUUAUAAAUCUCAAUUUAUGUCGGCUAUUUAGAAAUUUUGGGCACGCAGCAGCCAUUCCGACUCAGGG ------------------------..(((((((((...(((((..(((.(((((((....)))))))..)))..))))).........((.....)).)))))))))........ ( -29.50) >consensus _G____ACCAAGAA______CCAGGCCAUCAUGGCCACCUAAGCGCCCAUAUAAAUCUCAAUUUAUGUUGGCUAUUUAGAAAUUUCGGGCACGCAGCAGCCAUUUCGACUUAGGG .....................................((((((((.((((((((((....))))))).)))................(((........)))....)).)))))). (-12.55 = -13.47 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:08 2006