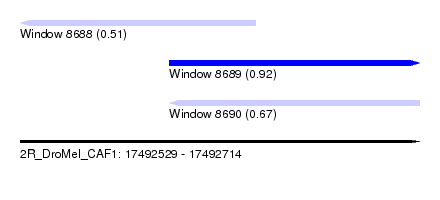
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,492,529 – 17,492,714 |
| Length | 185 |
| Max. P | 0.915591 |

| Location | 17,492,529 – 17,492,638 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 93.62 |
| Mean single sequence MFE | -20.38 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.99 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17492529 109 - 20766785 UAAUUAAGCGGUUUUAUUUGCGGCUUUGCCGCACAUUAAUUGGUCAUGUUAAAUGUAAAAUUUCAUUUGCGCAAAAGAAAAA-AAAAACGAAACGUUUCCAUUAAUGAAA ..................((((((...))))))(((((((.((..(((((..........((((.(((....))).))))..-........)))))..)))))))))... ( -20.35) >DroSim_CAF1 33248 110 - 1 UAAUUAAGCGGUUUUAUUUGCGGCUUUGCCGCACAUUAAUUGGUCAUGUUAAAUGUAAAAUUUCAUUUGCGCAAAAAAAAAAAAAAAACGAAACGUUUCCAUUAAUGAAA ..................((((((...))))))(((((((.((...(((((((((........)))))).)))............(((((...))))))))))))))... ( -20.20) >DroYak_CAF1 29357 100 - 1 UAAUUAAGCGGUUUUAUUUGCGGCUUUGCCGCACAUUAAUUGGUCAUGUUAAAUGUAAAAUUUCAUUUGCGCAAA----------AAACGAAACGUUUCCAUUAAUGAAA ..................((((((...))))))(((((((.((..((((((((((........))))).((....----------...)).)))))..)))))))))... ( -20.60) >consensus UAAUUAAGCGGUUUUAUUUGCGGCUUUGCCGCACAUUAAUUGGUCAUGUUAAAUGUAAAAUUUCAUUUGCGCAAAA_AAAAA_AAAAACGAAACGUUUCCAUUAAUGAAA ..................((((((...))))))(((((((.((...(((((((((........)))))).)))............(((((...))))))))))))))... (-20.10 = -20.10 + -0.00)

| Location | 17,492,598 – 17,492,714 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.13 |
| Mean single sequence MFE | -34.37 |
| Consensus MFE | -29.96 |
| Energy contribution | -29.96 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17492598 116 + 20766785 AUUAAUGUGCGGCAAAGCCGCAAAUAAAACCGCUUAAUUACGUGACUCGGCCAAAAGGCAGGCG---AACCA-GCAACAGCCAGCAACAAAGGCAAAGCGGCUGAAACGCAGAAAUUUGC .....(.((((....((((((.........(((((.....((.....))(((....))))))))---.....-......(((.........)))...))))))....)))).)....... ( -33.90) >DroSim_CAF1 33318 116 + 1 AUUAAUGUGCGGCAAAGCCGCAAAUAAAACCGCUUAAUUACGUGACUCGGCCAAAAGGCAGGCG---AACCA-GCAACAGCCAGCAACAAAGGCAAAGCGGCUGAAACGCAGAAAUUUGC .....(.((((....((((((.........(((((.....((.....))(((....))))))))---.....-......(((.........)))...))))))....)))).)....... ( -33.90) >DroYak_CAF1 29417 114 + 1 AUUAAUGUGCGGCAAAGCCGCAAAUAAAACCGCUUAAUUACGUGACUCGGCCAAAAGGCAGGCGGCGAACGGCGAACCAGCCAGCAACAAAGGCAAAGCGG------CGCAGAAAUUUGC .....(((((.((...(((..........((((((.....((.....))(((....))))))))).....(((......))).........)))...)).)------))))......... ( -35.30) >consensus AUUAAUGUGCGGCAAAGCCGCAAAUAAAACCGCUUAAUUACGUGACUCGGCCAAAAGGCAGGCG___AACCA_GCAACAGCCAGCAACAAAGGCAAAGCGGCUGAAACGCAGAAAUUUGC .......((((((...)))))).......((((((.....((.....))(((.....((.(((................))).))......))).)))))).......((((....)))) (-29.96 = -29.96 + -0.00)


| Location | 17,492,598 – 17,492,714 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.13 |
| Mean single sequence MFE | -37.43 |
| Consensus MFE | -31.07 |
| Energy contribution | -32.07 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.672630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17492598 116 - 20766785 GCAAAUUUCUGCGUUUCAGCCGCUUUGCCUUUGUUGCUGGCUGUUGC-UGGUU---CGCCUGCCUUUUGGCCGAGUCACGUAAUUAAGCGGUUUUAUUUGCGGCUUUGCCGCACAUUAAU (((......))).....((((((((.(((.........))).(((((-((..(---((...(((....))))))..)).))))).)))))))).....((((((...))))))....... ( -38.00) >DroSim_CAF1 33318 116 - 1 GCAAAUUUCUGCGUUUCAGCCGCUUUGCCUUUGUUGCUGGCUGUUGC-UGGUU---CGCCUGCCUUUUGGCCGAGUCACGUAAUUAAGCGGUUUUAUUUGCGGCUUUGCCGCACAUUAAU (((......))).....((((((((.(((.........))).(((((-((..(---((...(((....))))))..)).))))).)))))))).....((((((...))))))....... ( -38.00) >DroYak_CAF1 29417 114 - 1 GCAAAUUUCUGCG------CCGCUUUGCCUUUGUUGCUGGCUGGUUCGCCGUUCGCCGCCUGCCUUUUGGCCGAGUCACGUAAUUAAGCGGUUUUAUUUGCGGCUUUGCCGCACAUUAAU (((......)))(------((((((......(((.((((((((((..((.(....).))..)))....)))).))).))).....)))))))......((((((...))))))....... ( -36.30) >consensus GCAAAUUUCUGCGUUUCAGCCGCUUUGCCUUUGUUGCUGGCUGUUGC_UGGUU___CGCCUGCCUUUUGGCCGAGUCACGUAAUUAAGCGGUUUUAUUUGCGGCUUUGCCGCACAUUAAU (((......))).....((((((((.......((((((((((.......(((.....))).(((....)))..))))).))))).)))))))).....((((((...))))))....... (-31.07 = -32.07 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:05 2006