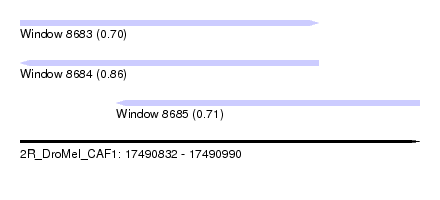
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,490,832 – 17,490,990 |
| Length | 158 |
| Max. P | 0.862128 |

| Location | 17,490,832 – 17,490,950 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.53 |
| Mean single sequence MFE | -28.01 |
| Consensus MFE | -23.44 |
| Energy contribution | -23.00 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17490832 118 + 20766785 AAACUGCUAAAUUUUUUCAUCCAUUUUUUUGGACC--CACACCAAUCGUUAGCAGCUGGUUGCAAUUGCACCGAUUUAGGUGAUAGCACCACCUUCGAGUGCACUUCAACUUUAGGGAUA ...(((((((.........((((......))))..--...........)))))))..(((((.((.((((((((...(((((.......)))))))).))))).)))))))......... ( -30.75) >DroSec_CAF1 18624 117 + 1 AAAUUGCUAAAUUUUUUCAUUCAUUU-UUUGUACC--CACUUCAACCGUUAGCAGCUGGUUGCAAUUGCACCGAUUUAGGUGAUAGCACCACCUUCGAGUGCACUUCAACUUUAGGGAUA ..(((.(((((...............-.(((((.(--((((.(........).)).))).))))).((((((((...(((((.......)))))))).))))).......))))).))). ( -26.50) >DroSim_CAF1 31524 117 + 1 AAACUGCUAAAUUUUUUCAUUCAUUU-UUUGUACC--CACACCAACCGUUAGCAGCUGGUUGCAAUUGCACCGAUUUAGGUGAUAGCACCACCUUCGAGUGCACUUCAACUUUAGGGAUA ..........................-......((--(....((((((........))))))....((((((((...(((((.......)))))))).)))))...........)))... ( -26.20) >DroYak_CAF1 27666 119 + 1 AAAGUACUCAACUUUUUCAUCCAUUU-UUUGGCCCCUCACACCAACCGUUAGCAGCUGGUUGCAAUUGCACCGAUUUAGGUGAUAGCACCACCUUCGAGUGCACUUCAAGUUUAGGGAUA (((((.....))))).....(((...-..))).((((.((..((((((........))))))....((((((((...(((((.......)))))))).)))))......))..))))... ( -28.60) >consensus AAACUGCUAAAUUUUUUCAUCCAUUU_UUUGGACC__CACACCAACCGUUAGCAGCUGGUUGCAAUUGCACCGAUUUAGGUGAUAGCACCACCUUCGAGUGCACUUCAACUUUAGGGAUA ..................((((......(((...........((((((........))))))....((((((((...(((((.......)))))))).)))))...)))......)))). (-23.44 = -23.00 + -0.44)

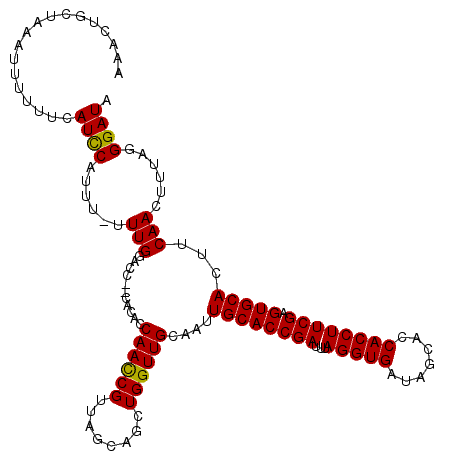
| Location | 17,490,832 – 17,490,950 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.53 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -27.67 |
| Energy contribution | -28.18 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17490832 118 - 20766785 UAUCCCUAAAGUUGAAGUGCACUCGAAGGUGGUGCUAUCACCUAAAUCGGUGCAAUUGCAACCAGCUGCUAACGAUUGGUGUG--GGUCCAAAAAAAUGGAUGAAAAAAUUUAGCAGUUU ..........((((...(((((.((((((((((...)))))))...))))))))....)))).((((((((((.((....)).--)(((((......)))))........))))))))). ( -35.90) >DroSec_CAF1 18624 117 - 1 UAUCCCUAAAGUUGAAGUGCACUCGAAGGUGGUGCUAUCACCUAAAUCGGUGCAAUUGCAACCAGCUGCUAACGGUUGAAGUG--GGUACAAA-AAAUGAAUGAAAAAAUUUAGCAAUUU (((((((...((((...(((((.((((((((((...)))))))...))))))))....))))((((((....)))))).)).)--))))....-...(((((......)))))....... ( -32.80) >DroSim_CAF1 31524 117 - 1 UAUCCCUAAAGUUGAAGUGCACUCGAAGGUGGUGCUAUCACCUAAAUCGGUGCAAUUGCAACCAGCUGCUAACGGUUGGUGUG--GGUACAAA-AAAUGAAUGAAAAAAUUUAGCAGUUU ..........((((((.(((((.((((((((((...)))))))...)))))))).(..((.(((((((....)))))))))..--).......-...............))))))..... ( -33.50) >DroYak_CAF1 27666 119 - 1 UAUCCCUAAACUUGAAGUGCACUCGAAGGUGGUGCUAUCACCUAAAUCGGUGCAAUUGCAACCAGCUGCUAACGGUUGGUGUGAGGGGCCAAA-AAAUGGAUGAAAAAGUUGAGUACUUU ..((((...........(((((.((((((((((...)))))))...)))))))).(..((.(((((((....)))))))))..)))))(((..-...))).....(((((.....))))) ( -37.00) >consensus UAUCCCUAAAGUUGAAGUGCACUCGAAGGUGGUGCUAUCACCUAAAUCGGUGCAAUUGCAACCAGCUGCUAACGGUUGGUGUG__GGUACAAA_AAAUGAAUGAAAAAAUUUAGCAGUUU .................(((((.((((((((((...)))))))...))))))))(((((.((((((((....)))))))).................(((((......)))))))))).. (-27.67 = -28.18 + 0.50)

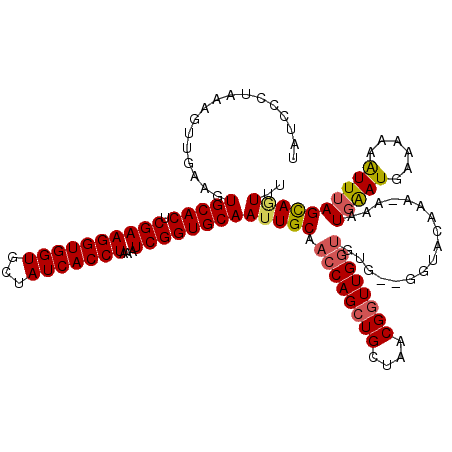
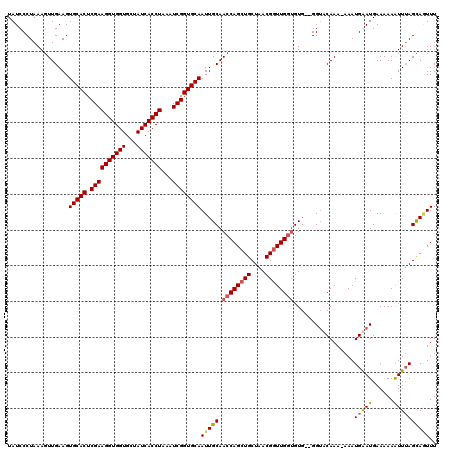
| Location | 17,490,870 – 17,490,990 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.92 |
| Mean single sequence MFE | -36.91 |
| Consensus MFE | -34.62 |
| Energy contribution | -35.37 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
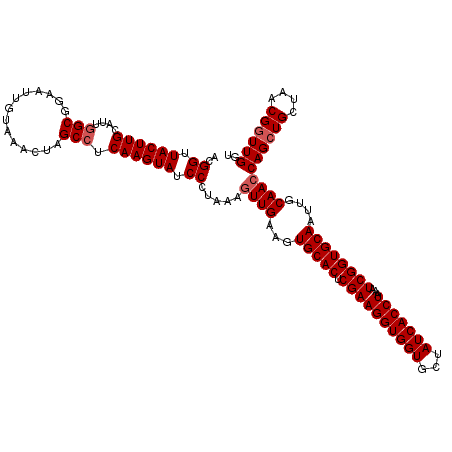
>2R_DroMel_CAF1 17490870 120 - 20766785 ACGGUUACUUGCAUUGGCGGAAUUGUAAACUAGCAUCAAGUAUCCCUAAAGUUGAAGUGCACUCGAAGGUGGUGCUAUCACCUAAAUCGGUGCAAUUGCAACCAGCUGCUAACGAUUGGU ......(((..(.(((((((..((((((....((.((((.(........).)))).))((((.((((((((((...)))))))...)))))))..))))))....))))))).)...))) ( -31.90) >DroSec_CAF1 18661 120 - 1 ACGGUUACUUGCAUUGGCGGAAUUGUAAACUAGCCUCAAGUAUCCCUAAAGUUGAAGUGCACUCGAAGGUGGUGCUAUCACCUAAAUCGGUGCAAUUGCAACCAGCUGCUAACGGUUGAA ..((.((((((....(((..............))).)))))).)).....((((...(((((.((((((((((...)))))))...))))))))....))))((((((....)))))).. ( -38.34) >DroSim_CAF1 31561 120 - 1 ACGGUUACUUGCAUUGGCGGAAUUGUAAACUAGCCUCAAGUAUCCCUAAAGUUGAAGUGCACUCGAAGGUGGUGCUAUCACCUAAAUCGGUGCAAUUGCAACCAGCUGCUAACGGUUGGU ..((.((((((....(((..............))).)))))).)).....((.....(((((.((((((((((...)))))))...))))))))...)).((((((((....)))))))) ( -38.74) >DroYak_CAF1 27705 120 - 1 ACGGUUACUUGCAUUGGCGGAAUUGUAAACUAGCCUCAAGUAUCCCUAAACUUGAAGUGCACUCGAAGGUGGUGCUAUCACCUAAAUCGGUGCAAUUGCAACCAGCUGCUAACGGUUGGU ..((.((((((....(((..............))).)))))).)).......((...(((((.((((((((((...)))))))...))))))))....))((((((((....)))))))) ( -38.64) >consensus ACGGUUACUUGCAUUGGCGGAAUUGUAAACUAGCCUCAAGUAUCCCUAAAGUUGAAGUGCACUCGAAGGUGGUGCUAUCACCUAAAUCGGUGCAAUUGCAACCAGCUGCUAACGGUUGGU ..((.((((((....(((..............))).)))))).)).....((((...(((((.((((((((((...)))))))...))))))))....))))((((((....)))))).. (-34.62 = -35.37 + 0.75)

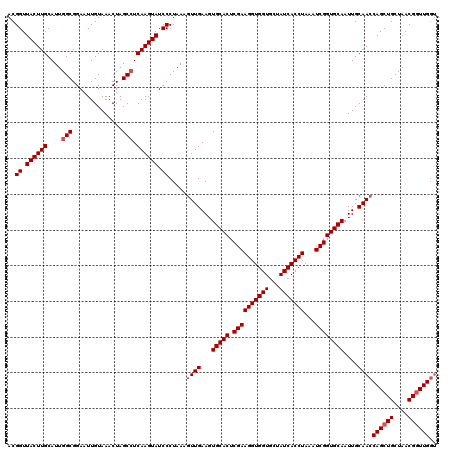
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:00 2006