| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,862,013 – 2,862,234 |
| Length | 221 |
| Max. P | 0.745862 |

| Location | 2,862,013 – 2,862,133 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.55 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -18.74 |
| Energy contribution | -20.77 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
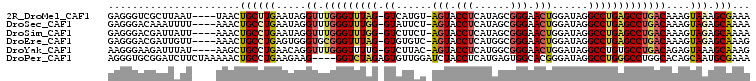
>2R_DroMel_CAF1 2862013 120 - 20766785 AAAACAAUCUUAAGUUUCCAAAUUUAUUCGAACUUUCAGAGCCCGUGGGCUCCAAGGCUCCAACCUUUGCUACGGCUUCAAAGAUUUCCAGUCUACUCGGCUCGAGUUCGAGUGAUAUUG ..................(((..((((((((((((...(((((.(((((((..((((......)))).((....)).............)))))))..)))))))))))))))))..))) ( -36.10) >DroSec_CAF1 13646 120 - 1 AAAACACUCCUAAGUUUCCAAAUUUGUUCGAACUUUCAGAGCCCGUGGGCUCCAAGGCUCCAACCUUUGCUACGGCUUCAAAGAUUUCCAGUCUACUCGGCUCGAGUUCGAGUAAUAUAG .......................((((((((((((...(((((.(((((((..((((......)))).((....)).............)))))))..)))))))))))))))))..... ( -32.70) >DroSim_CAF1 20341 120 - 1 AAAACCAUCCUAAGUUUCAAAAUUGAUUUGAACUUUCAGAGCCCGUGGGCUCCAAGGCUCCAACCUUUGCUACGGCUUCUAAGAUUUCCAGUCUACUCGGCUCGAGUUCGAGUGAUAUAG .......................(.((((((((((...(((((.(((((((..((((......)))).((....)).............)))))))..))))))))))))))).)..... ( -31.80) >DroEre_CAF1 13240 114 - 1 AAAACUCUCCUAAGU-CUUGA-----UUCCAACUUUCAGAGCCCGUGGGCUCCAAAGCUCCCACCUUUGCUACGGCUUCAAAGAUUUCCAGCACACUGGGCUCAAGCUCGAGCGAUAUUG ...........((((-(((((-----..((........(((((....)))))(((((.......)))))....))..)).)))))))((((....))))((((......))))....... ( -29.80) >DroYak_CAF1 15459 119 - 1 AAAACACUCCCAGUU-CUUAAAUUUAUUUAAUGUUCCAGAGCCCGUGGGCUCCAAGGCUCCAACCUUUGCUACGGCUUCAAAGAUUUCAAGUAUGCUCGACUCAAGUUCGAGUGAUAUUG ....(((((..(((.-(((((((((.............(((((....))))).((((......)))).((....)).....)))))).)))...))).(((....))).)))))...... ( -24.40) >DroPer_CAF1 13691 103 - 1 AAAAGUA----------------UUGUUUG-AUUUGCAGAACCCGUUGGGGCCAAGGCGCCCACAUUUGCCAGUGUCUCGAAAAUAUCCAGUCUACUCGGCUCCAGUUCGAGUGAUAUUG ...((((----------------(..((((-(.(((........((.(((((....)).)))))....(((((((.((.((.....)).)).).))).)))..))).)))))..))))). ( -27.10) >consensus AAAACAAUCCUAAGU_UCCAAAUUUAUUCGAACUUUCAGAGCCCGUGGGCUCCAAGGCUCCAACCUUUGCUACGGCUUCAAAGAUUUCCAGUCUACUCGGCUCGAGUUCGAGUGAUAUUG .......................((((((((((((...(((((.(((((((..((((......)))).((....)).............)))))))..)))))))))))))))))..... (-18.74 = -20.77 + 2.03)


| Location | 2,862,053 – 2,862,165 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 72.96 |
| Mean single sequence MFE | -31.29 |
| Consensus MFE | -16.85 |
| Energy contribution | -17.00 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2862053 112 + 20766785 UGAAGCCGUAGCAAAGGUUGGAGCCUUGGAGCCCACGGGCUCUGAAAGUUCGAAUAAAUUUGGAAACUUAAGAUUGUUUUGAGG-GUCGCUUAAU----UAACUGCUUGAAUAGGUU (.((((.((..((((.(((.((((.(..(((((....)))))..)..)))).)))...))))((..(((((((...))))))).-.)).......----..)).)))).)....... ( -32.20) >DroGri_CAF1 14303 110 + 1 UGAGACACUAGCAAAUGUGGGCGCUUUGGCACCGACGGGUUCUGGAAUUUG-AGUG-AGAGAU--AGUCACAAUUCGUUAGAAAACUAUA--UAAUUGAUAACUGUCUGAAU-GUUA ...((((((((.(..(((.((.((....)).)).)))..).))))......-....-..((((--(((..(((((...(((....)))..--.)))))...)))))))...)-))). ( -25.10) >DroSec_CAF1 13686 112 + 1 UGAAGCCGUAGCAAAGGUUGGAGCCUUGGAGCCCACGGGCUCUGAAAGUUCGAACAAAUUUGGAAACUUAGGAGUGUUUUGAGG-GACAAAUUUU----AAACUGCCUGAAUAGGUU ...((((....((((.(((.((((.(..(((((....)))))..)..)))).)))...)))).....(((((...((((.(((.-......))).----))))..)))))...)))) ( -33.00) >DroSim_CAF1 20381 112 + 1 AGAAGCCGUAGCAAAGGUUGGAGCCUUGGAGCCCACGGGCUCUGAAAGUUCAAAUCAAUUUUGAAACUUAGGAUGGUUUUGAGG-GACGAUUAUU----AAACUGCCUGAAUAGGUU ...((((.......((((.(((((.(..(((((....)))))..)..)))).((((..(((..(((((......)))))..)))-...))))...----...).)))).....)))) ( -32.60) >DroEre_CAF1 13280 106 + 1 UGAAGCCGUAGCAAAGGUGGGAGCUUUGGAGCCCACGGGCUCUGAAAGUUGGAA-----UCAAG-ACUUAGGAGAGUUUUGAGG-GACGAUUGUU----AAACUGCCUGAGUGGGUG ....(((...((..(((..(.(((((..(((((....)))))..))(((((...-----(((((-((((....)))))))))..-..))))))))----...)..)))..)).))). ( -38.90) >DroYak_CAF1 15499 111 + 1 UGAAGCCGUAGCAAAGGUUGGAGCCUUGGAGCCCACGGGCUCUGGAACAUUAAAUAAAUUUAAG-AACUGGGAGUGUUUUAAGG-GAAGAUUUAU----AAGCUGCCUGAACAGGUU ...(((((((((.(((((....)))))((((((....)))))).......(((((...(((.((-(((.......))))).)))-....))))).----..))))).......)))) ( -25.91) >consensus UGAAGCCGUAGCAAAGGUUGGAGCCUUGGAGCCCACGGGCUCUGAAAGUUCGAAUAAAUUUAGA_ACUUAGGAGUGUUUUGAGG_GACGAUUUAU____AAACUGCCUGAAUAGGUU ....(((.......(((..(.....(..(((((....)))))..).....................(((((((...)))))))...................)..))).....))). (-16.85 = -17.00 + 0.15)

| Location | 2,862,053 – 2,862,165 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.96 |
| Mean single sequence MFE | -20.45 |
| Consensus MFE | -13.70 |
| Energy contribution | -12.82 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2862053 112 - 20766785 AACCUAUUCAAGCAGUUA----AUUAAGCGAC-CCUCAAAACAAUCUUAAGUUUCCAAAUUUAUUCGAACUUUCAGAGCCCGUGGGCUCCAAGGCUCCAACCUUUGCUACGGCUUCA .........((((.((..----....(((((.-..............((((((....))))))...((.((((..(((((....))))).)))).))......))))))).)))).. ( -18.10) >DroGri_CAF1 14303 110 - 1 UAAC-AUUCAGACAGUUAUCAAUUA--UAUAGUUUUCUAACGAAUUGUGACU--AUCUCU-CACU-CAAAUUCCAGAACCCGUCGGUGCCAAAGCGCCCACAUUUGCUAGUGUCUCA ....-....(((((.(((.(((...--....(..((((...(((((((((..--.....)-))).-..))))).))))..)((.(((((....))))).))..))).)))))))).. ( -20.80) >DroSec_CAF1 13686 112 - 1 AACCUAUUCAGGCAGUUU----AAAAUUUGUC-CCUCAAAACACUCCUAAGUUUCCAAAUUUGUUCGAACUUUCAGAGCCCGUGGGCUCCAAGGCUCCAACCUUUGCUACGGCUUCA ..(((.....((.(((((----.(((((((..-.....((((........)))).)))))))....))))).)).(((((....)))))..)))...........((....)).... ( -20.30) >DroSim_CAF1 20381 112 - 1 AACCUAUUCAGGCAGUUU----AAUAAUCGUC-CCUCAAAACCAUCCUAAGUUUCAAAAUUGAUUUGAACUUUCAGAGCCCGUGGGCUCCAAGGCUCCAACCUUUGCUACGGCUUCU ..(((.....((.(((((----((..((((..-.....((((........))))......))))))))))).)).(((((....)))))..)))...........((....)).... ( -21.62) >DroEre_CAF1 13280 106 - 1 CACCCACUCAGGCAGUUU----AACAAUCGUC-CCUCAAAACUCUCCUAAGU-CUUGA-----UUCCAACUUUCAGAGCCCGUGGGCUCCAAAGCUCCCACCUUUGCUACGGCUUCA ..........((.(((((----..........-..((((.(((......)))-.))))-----............(((((....)))))..))))).))......((....)).... ( -21.90) >DroYak_CAF1 15499 111 - 1 AACCUGUUCAGGCAGCUU----AUAAAUCUUC-CCUUAAAACACUCCCAGUU-CUUAAAUUUAUUUAAUGUUCCAGAGCCCGUGGGCUCCAAGGCUCCAACCUUUGCUACGGCUUCA ..........((.(((((----..........-..(((((((.......)))-.)))).................(((((....)))))..))))))).......((....)).... ( -20.00) >consensus AACCUAUUCAGGCAGUUU____AAAAAUCGUC_CCUCAAAACAAUCCUAAGU_UCUAAAUUUAUUCGAACUUUCAGAGCCCGUGGGCUCCAAGGCUCCAACCUUUGCUACGGCUUCA .........((((..............................................................(((((....)))))...(((..........)))...)))).. (-13.70 = -12.82 + -0.89)

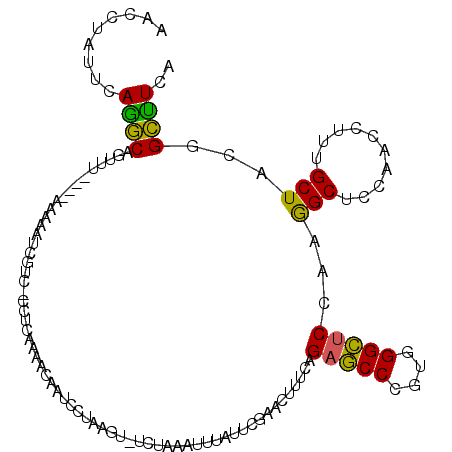
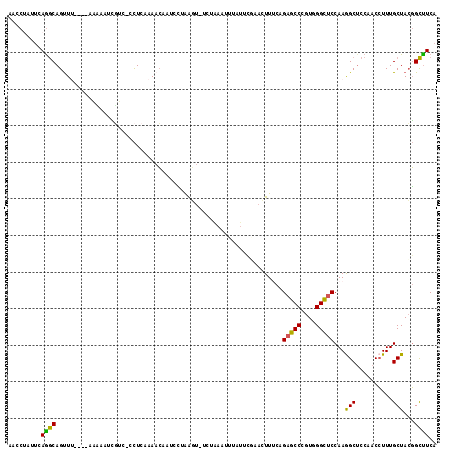
| Location | 2,862,133 – 2,862,234 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -32.14 |
| Consensus MFE | -16.55 |
| Energy contribution | -16.75 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2862133 101 + 20766785 GAGGGUCGCUUAAU----UAACUGCUUGAAUAGGUUUGGGUUUAG-GUCAUGU-AGUACCUCAUAGCGGGAACUGGAUAGGCCUGAGCCUGACAAAGUAAAGCGAAA .....((((((...----....(((((......(.(..(((((((-(((((.(-(((.(((......))).)))).)).))))))))))..)).))))))))))).. ( -33.51) >DroSec_CAF1 13766 101 + 1 GAGGGACAAAUUUU----AAACUGCCUGAAUAGGUUUGGGUUUGG-GUAUUCU-AGUACCUCAUAGCGGGAACUGGAUAGGCCUGAGCCUGACAAAGUAGAGCAAAA ..............----....((((((.....(.(..((((..(-((..(((-(((.(((......))).))))))...)))..))))..))....))).)))... ( -31.60) >DroSim_CAF1 20461 101 + 1 GAGGGACGAUUAUU----AAACUGCCUGAAUAGGUUUGGGUUUGG-GUCUUCU-AGUACCUCAUAGCGGGAACUGGAUAGGCCUGAGCCUGACAAAGUAGAGCAAAA ..............----....((((((.....(.(..((((..(-(((((((-(((.(((......))).)))))).)))))..))))..))....))).)))... ( -34.20) >DroEre_CAF1 13354 101 + 1 GAGGGACGAUUGUU----AAACUGCCUGAGUGGGUGCGGGUUUAG-GUGUGUC-AGUACCUCAUGGCGGGAACUGGAUAGGCCUGAGCCUGACAAAGUAGAGCAAAG .........(((((----..(((((((....)))).(((((((((-((.((((-(((.(((......))).))).)))).)))))))))))....)))..))))).. ( -36.80) >DroYak_CAF1 15578 101 + 1 AAGGGAAGAUUUAU----AAGCUGCCUGAACAGGUUUGGGUUUUG-GUCUUAC-AGUACCUCAUGGCGGGAACUGGAUAGGCCUGUGCCUGACAGAGUAAAGCAAAG ..............----..(((((((....))))(..(((...(-((((..(-(((.(((......))).))))...)))))...)))..)........))).... ( -28.40) >DroPer_CAF1 13794 103 + 1 AGGGUGCGGAUCUUCUAAAAACUGCCUGAAGAAG----GGUCUAGAGUGUUGGAUCUACCUCAUGAGUGGCACGGGAUAGGCCUGGGCCUGGCACAGCAAUGCGAAA ...((((((.(((((............))))).(----(((((((....)))))))).))........(((.((((.....)))).)))..))))............ ( -28.30) >consensus GAGGGACGAUUAUU____AAACUGCCUGAAUAGGUUUGGGUUUAG_GUCUUCU_AGUACCUCAUAGCGGGAACUGGAUAGGCCUGAGCCUGACAAAGUAAAGCAAAA ......................((((((.....((.(((((((((.((......(((.(((......))).)))......)))))))))))))....))).)))... (-16.55 = -16.75 + 0.20)

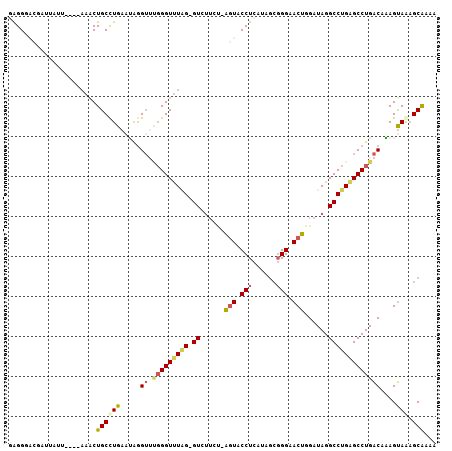
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:23 2006