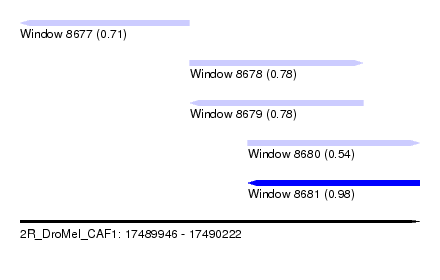
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,489,946 – 17,490,222 |
| Length | 276 |
| Max. P | 0.978534 |

| Location | 17,489,946 – 17,490,063 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.78 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -24.49 |
| Energy contribution | -24.55 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17489946 117 - 20766785 GACCCACAUUUGUGUGGUUCCCCU-AUUCUGGACUCGCCC--ACCCGCUUUUCCGGGUGUUGAUAACUGUUUUUCCGACCUCCAAAUUCCCCGCAAACUCCUCGGAAAACUUGUCCAUGU ((.(((((....))))).))....-....((((((((..(--(((((......)))))).))).......((((((((.......................))))))))...)))))... ( -29.40) >DroSec_CAF1 17772 117 - 1 GACCCACAUUUGUGUGGUUCCCCC-AUUCUGGACUCGCCC--ACCCGCUUUUCCGGGUGUUGAUAACUGUUUUUCCGACCUCCAAAUUCCCCGCAAACUCCUCGGAAAACUUGUCCAUGU ((.(((((....))))).))....-....((((((((..(--(((((......)))))).))).......((((((((.......................))))))))...)))))... ( -29.40) >DroSim_CAF1 30634 117 - 1 GACCCACAUUUGUGUGGUUCCCCC-AUUCUGGACUCGCCC--ACCCGCUUUUCCGUGUGUUGAUAACUGUUUUUCCGACCUCCAAAUUCCCCGCAAACUCCUCGGAAAACUUGUCCAUGU ((.(((((....))))).))....-....((((((((..(--((.((......)).))).))).......((((((((.......................))))))))...)))))... ( -22.30) >DroYak_CAF1 26808 120 - 1 GACCCACAUUUGUGUGGUUCCCCCCUUUUUGGAUGCGCCCACACCCGCUUUUCCGGGUGUUGAUAACUGUUUUUCCGACCUCCAAAUCCCCCGCAAACUCCUCGGAAAACUUGUCCAUGU ((.(((((....))))).)).........(((((....(.(((((((......))))))).)........((((((((.......................))))))))...)))))... ( -29.40) >consensus GACCCACAUUUGUGUGGUUCCCCC_AUUCUGGACUCGCCC__ACCCGCUUUUCCGGGUGUUGAUAACUGUUUUUCCGACCUCCAAAUUCCCCGCAAACUCCUCGGAAAACUUGUCCAUGU ((.(((((....))))).)).........(((((........(((((......)))))............((((((((.......................))))))))...)))))... (-24.49 = -24.55 + 0.06)

| Location | 17,490,063 – 17,490,183 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -22.20 |
| Energy contribution | -23.76 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17490063 120 + 20766785 AAGUUAUUUUCAACACUUAAAAUAAACUCUGCCUCCUCGGCUCGGUUCCUUGCUCUUUGCCAGAAUCCUGGCCAGCAUUUCUUUAGCCGCCCAUUUUCUGGGUCAGUCGCAAACACAGCC ...(((((((........)))))))....(((..(...((((........((((....(((((....))))).)))).......))))(((((.....)))))..)..)))......... ( -26.16) >DroSec_CAF1 17889 120 + 1 AAGUUAUUUUCAACACUUAAAAUAAACUCAGCUGCCUCGGCUCGGUUCCCUGCUCUUUGCCAGAAUCCUGGCCAGCAUUUCUUUAGCCGCCCAUUUUCUGGGUCAGUCGCAAACACAGCC ...(((((((........))))))).....((((...(((((.((((...((((....(((((....))))).)))).......))))(((((.....))))).)))))......)))). ( -28.10) >DroSim_CAF1 30751 120 + 1 AAGUUAUUUUCAACACUUAAAAUAAACUCAGCUGCCUCGGCUCGGUUCCUUGCUCUUUGCCAGAAUCCUGGCCAGCAUUUCUUUAGCCGCCCAUUUUCUGGGUCAGUCGCAAACACAGCC ...(((((((........))))))).....((((...(((((.((((...((((....(((((....))))).)))).......))))(((((.....))))).)))))......)))). ( -28.80) >DroYak_CAF1 26928 106 + 1 AAGUUAUUUUCAACACUUAAAAUAAACUCUGCUGCCUCGGC--------------UUUGCCAGAAUCCUGGCCAGCAUUUCUUUAGCCGCCCAUUUUCUGGGUCAGUCGCAAGCACAGUC ...(((((((........)))))))(((.((((((..((((--------------(..(((((....)))))............))))(((((.....)))))..)..)).)))).))). ( -27.64) >consensus AAGUUAUUUUCAACACUUAAAAUAAACUCAGCUGCCUCGGCUCGGUUCCUUGCUCUUUGCCAGAAUCCUGGCCAGCAUUUCUUUAGCCGCCCAUUUUCUGGGUCAGUCGCAAACACAGCC ...(((((((........))))))).....((((....((((........((((....(((((....))))).)))).......))))(((((.....)))))............)))). (-22.20 = -23.76 + 1.56)


| Location | 17,490,063 – 17,490,183 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -28.47 |
| Energy contribution | -30.03 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.776784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17490063 120 - 20766785 GGCUGUGUUUGCGACUGACCCAGAAAAUGGGCGGCUAAAGAAAUGCUGGCCAGGAUUCUGGCAAAGAGCAAGGAACCGAGCCGAGGAGGCAGAGUUUAUUUUAAGUGUUGAAAAUAACUU ((...(.(((((..((...((((((..(((.((((.........)))).)))...))))))...)).))))).).))..(((.....))).(((((.(((((........)))))))))) ( -35.40) >DroSec_CAF1 17889 120 - 1 GGCUGUGUUUGCGACUGACCCAGAAAAUGGGCGGCUAAAGAAAUGCUGGCCAGGAUUCUGGCAAAGAGCAGGGAACCGAGCCGAGGCAGCUGAGUUUAUUUUAAGUGUUGAAAAUAACUU ((((((............((((.....))))(((((.......((((.(((((....)))))....))))........)))))..))))))(((((.(((((........)))))))))) ( -37.06) >DroSim_CAF1 30751 120 - 1 GGCUGUGUUUGCGACUGACCCAGAAAAUGGGCGGCUAAAGAAAUGCUGGCCAGGAUUCUGGCAAAGAGCAAGGAACCGAGCCGAGGCAGCUGAGUUUAUUUUAAGUGUUGAAAAUAACUU ((((((.((.(((.(((.((((.....))))))))........((((.(((((....)))))....)))).........)).)).))))))(((((.(((((........)))))))))) ( -36.40) >DroYak_CAF1 26928 106 - 1 GACUGUGCUUGCGACUGACCCAGAAAAUGGGCGGCUAAAGAAAUGCUGGCCAGGAUUCUGGCAAA--------------GCCGAGGCAGCAGAGUUUAUUUUAAGUGUUGAAAAUAACUU ..((((((((.((.((...((((((..(((.((((.........)))).)))...))))))...)--------------).))))))).)))((((.(((((........))))))))). ( -33.20) >consensus GGCUGUGUUUGCGACUGACCCAGAAAAUGGGCGGCUAAAGAAAUGCUGGCCAGGAUUCUGGCAAAGAGCAAGGAACCGAGCCGAGGCAGCAGAGUUUAUUUUAAGUGUUGAAAAUAACUU .(((((............((((.....))))(((((.......((((.(((((....)))))....))))........)))))..))))).(((((.(((((........)))))))))) (-28.47 = -30.03 + 1.56)

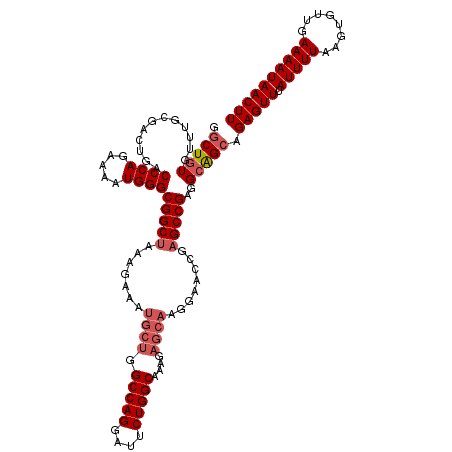
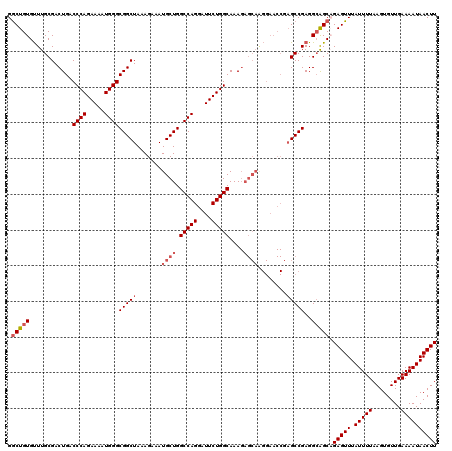
| Location | 17,490,103 – 17,490,222 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.74 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -17.66 |
| Energy contribution | -18.98 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17490103 119 + 20766785 CUCGGUUCCUUGCUCUUUGCCAGAAUCCUGGCCAGCAUUUCUUUAGCCGCCCAUUUUCUGGGUCAGUCGCAAACACAGCCCCCCCUUUUCCGACCA-UUUUCCAACCGAAACCCUCUUUG .((((((...((((....(((((....))))).))))...........(((((.....)))))..((((.(((.............))).))))..-......))))))........... ( -25.12) >DroSec_CAF1 17929 118 + 1 CUCGGUUCCCUGCUCUUUGCCAGAAUCCUGGCCAGCAUUUCUUUAGCCGCCCAUUUUCUGGGUCAGUCGCAAACACAGCCC-CCCUUUUCCGACCA-UUUUCCAACCGAAACCCUCUUUG .((((((...((((....(((((....))))).))))...........(((((.....)))))..((((.(((........-....))).))))..-......))))))........... ( -25.10) >DroSim_CAF1 30791 118 + 1 CUCGGUUCCUUGCUCUUUGCCAGAAUCCUGGCCAGCAUUUCUUUAGCCGCCCAUUUUCUGGGUCAGUCGCAAACACAGCCC-CCCUUUUCCGACCA-UUUUCCAACCGAAACCCUCUUUG .((((((...((((....(((((....))))).))))...........(((((.....)))))..((((.(((........-....))).))))..-......))))))........... ( -25.20) >DroYak_CAF1 26968 105 + 1 C--------------UUUGCCAGAAUCCUGGCCAGCAUUUCUUUAGCCGCCCAUUUUCUGGGUCAGUCGCAAGCACAGUCC-CGCUUUUCCGACCACUUUUCCAACCGAAACCCUCUUUG .--------------...(((((....))))).((..((((.......(((((.....)))))..((((.((((.......-.))))...)))).............))))..))..... ( -21.80) >consensus CUCGGUUCCUUGCUCUUUGCCAGAAUCCUGGCCAGCAUUUCUUUAGCCGCCCAUUUUCUGGGUCAGUCGCAAACACAGCCC_CCCUUUUCCGACCA_UUUUCCAACCGAAACCCUCUUUG .((((((...........(((((....)))))................(((((.....)))))..((((.(((...((......))))).)))).........))))))........... (-17.66 = -18.98 + 1.31)

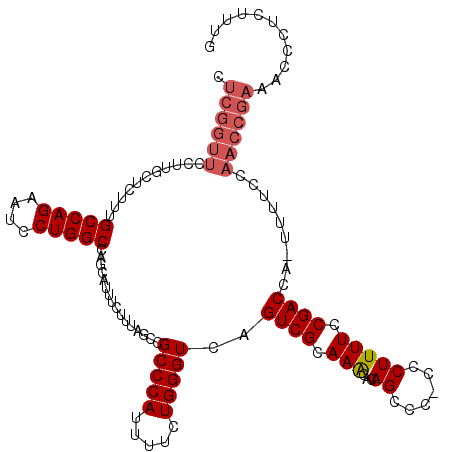
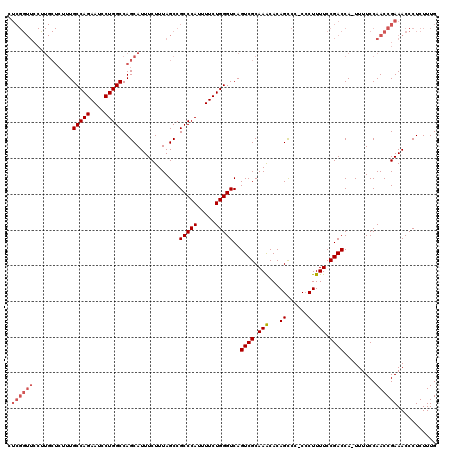
| Location | 17,490,103 – 17,490,222 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.74 |
| Mean single sequence MFE | -36.17 |
| Consensus MFE | -31.26 |
| Energy contribution | -32.33 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17490103 119 - 20766785 CAAAGAGGGUUUCGGUUGGAAAA-UGGUCGGAAAAGGGGGGGCUGUGUUUGCGACUGACCCAGAAAAUGGGCGGCUAAAGAAAUGCUGGCCAGGAUUCUGGCAAAGAGCAAGGAACCGAG .......(((((((((((.....-.(((((.(((.((.....))...))).)))))..((((.....))))))))).......((((.(((((....)))))....)))).))))))... ( -37.30) >DroSec_CAF1 17929 118 - 1 CAAAGAGGGUUUCGGUUGGAAAA-UGGUCGGAAAAGGG-GGGCUGUGUUUGCGACUGACCCAGAAAAUGGGCGGCUAAAGAAAUGCUGGCCAGGAUUCUGGCAAAGAGCAGGGAACCGAG .......(((((((((((.....-.(((((.(((.((.-...))...))).)))))..((((.....))))))))).......((((.(((((....)))))....)))).))))))... ( -37.10) >DroSim_CAF1 30791 118 - 1 CAAAGAGGGUUUCGGUUGGAAAA-UGGUCGGAAAAGGG-GGGCUGUGUUUGCGACUGACCCAGAAAAUGGGCGGCUAAAGAAAUGCUGGCCAGGAUUCUGGCAAAGAGCAAGGAACCGAG .......(((((((((((.....-.(((((.(((.((.-...))...))).)))))..((((.....))))))))).......((((.(((((....)))))....)))).))))))... ( -37.30) >DroYak_CAF1 26968 105 - 1 CAAAGAGGGUUUCGGUUGGAAAAGUGGUCGGAAAAGCG-GGACUGUGCUUGCGACUGACCCAGAAAAUGGGCGGCUAAAGAAAUGCUGGCCAGGAUUCUGGCAAA--------------G ...........(((((((.((..(..(((.........-.)))..)..)).))))))).((((((..(((.((((.........)))).)))...))))))....--------------. ( -33.00) >consensus CAAAGAGGGUUUCGGUUGGAAAA_UGGUCGGAAAAGGG_GGGCUGUGUUUGCGACUGACCCAGAAAAUGGGCGGCUAAAGAAAUGCUGGCCAGGAUUCUGGCAAAGAGCAAGGAACCGAG .......(((((((((((.(((...((((...........))))...))).))))))).((((((..(((.((((.........)))).)))...))))))............))))... (-31.26 = -32.33 + 1.06)

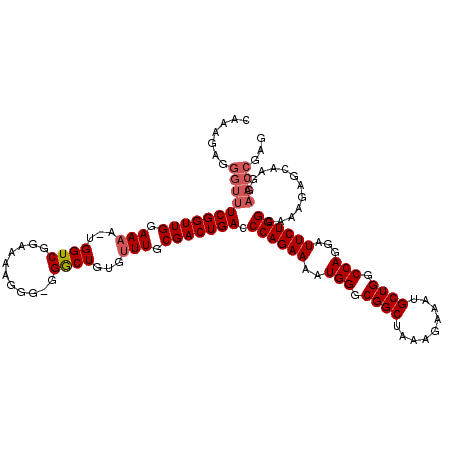

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:56 2006