| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,487,975 – 17,488,135 |
| Length | 160 |
| Max. P | 0.998562 |

| Location | 17,487,975 – 17,488,095 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.97 |
| Mean single sequence MFE | -31.05 |
| Consensus MFE | -22.45 |
| Energy contribution | -23.70 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17487975 120 + 20766785 AUGAUGACGGGAAUAUGUGUGAUCCUAUUAGUUCUCCACUGGCAAAGUGAGAGACUUCAAAUAGCACUCAAGGACUUGUGCUCCUUUCUUAAUCAUGAAGAACGCAACUCCUUGACUUUA ..(((.(((........))).)))((((((((.(((((((.....)))).))))))...)))))...(((((((.(((((.((..(((........))))).))))).)))))))..... ( -30.20) >DroSec_CAF1 16076 120 + 1 AUGAUGACGGGAAUAUGAGUUAUCCUGUUUGUUCUCCACUGGCAAAGUGAGAGGCUUCAAAUAGCACUCAAGGACUUGUGCUCCUUUCUUAAUUAUGAAGAGCGCAACUCCUUAACUUUG ..((.(((((((.((.....)))))))))...))........((((((..((((((......))).)))(((((.((((((((..(((........))))))))))).))))).)))))) ( -37.50) >DroSim_CAF1 28393 120 + 1 NNNNNNNNNNNNNNNNNNNAUUCCNUGUUCGUUCUCCACUGGCAAAGUGAGAGGCUUCAAAUAGCGCUCAAGGACUUGUGCUCCUUUCUCAAUUAUGAAGAGCGCAACUCCUUAACUUUG ..........................................((((((..((((((......))).)))(((((.((((((((..(((........))))))))))).))))).)))))) ( -29.80) >DroYak_CAF1 24288 120 + 1 AUGAUGACUGGAAUAUGUGUGAUCCUGCUAAUUCUCCACUGGCAGAGUGAGAGGCUUCAUAUAGCACUCAAGGAUCUGUGCUCCUUUCUAAAUUAUGAAGAACGCAACGCCUUAGUUUUA .....((((((.....((((((((((....(((((((...)).)))))..((((((......))).))).))))))((((.((..(((........))))).)))))))).))))))... ( -26.70) >consensus AUGAUGACGGGAAUAUGUGUGAUCCUGUUAGUUCUCCACUGGCAAAGUGAGAGGCUUCAAAUAGCACUCAAGGACUUGUGCUCCUUUCUUAAUUAUGAAGAACGCAACUCCUUAACUUUA .................................(((((((.....)))).)))(((......)))....(((((.((((((((..(((........))))))))))).)))))....... (-22.45 = -23.70 + 1.25)


| Location | 17,487,975 – 17,488,095 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.97 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -24.94 |
| Energy contribution | -24.62 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.710991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17487975 120 - 20766785 UAAAGUCAAGGAGUUGCGUUCUUCAUGAUUAAGAAAGGAGCACAAGUCCUUGAGUGCUAUUUGAAGUCUCUCACUUUGCCAGUGGAGAACUAAUAGGAUCACACAUAUUCCCGUCAUCAU .....(((((((.(((.(((((((........))..))))).))).)))))))((((((((.....((((.((((.....))))))))...)))))...))).................. ( -32.40) >DroSec_CAF1 16076 120 - 1 CAAAGUUAAGGAGUUGCGCUCUUCAUAAUUAAGAAAGGAGCACAAGUCCUUGAGUGCUAUUUGAAGCCUCUCACUUUGCCAGUGGAGAACAAACAGGAUAACUCAUAUUCCCGUCAUCAU ...(.(((((((.(((.(((((((........))..))))).))).))))))).)(((......)))(((.((((.....))))))).....((.(((..........))).))...... ( -29.90) >DroSim_CAF1 28393 120 - 1 CAAAGUUAAGGAGUUGCGCUCUUCAUAAUUGAGAAAGGAGCACAAGUCCUUGAGCGCUAUUUGAAGCCUCUCACUUUGCCAGUGGAGAACGAACANGGAAUNNNNNNNNNNNNNNNNNNN .....(((((((.(((.((((((((....)))....))))).))).)))))))..(((......)))(((.((((.....)))))))................................. ( -28.70) >DroYak_CAF1 24288 120 - 1 UAAAACUAAGGCGUUGCGUUCUUCAUAAUUUAGAAAGGAGCACAGAUCCUUGAGUGCUAUAUGAAGCCUCUCACUCUGCCAGUGGAGAAUUAGCAGGAUCACACAUAUUCCAGUCAUCAU .........(((((((.(((((((((....(((((((((.......)))))(((.(((......))))))....))))...))))))))))))).(((..........))).)))..... ( -28.70) >consensus CAAAGUUAAGGAGUUGCGCUCUUCAUAAUUAAGAAAGGAGCACAAGUCCUUGAGUGCUAUUUGAAGCCUCUCACUUUGCCAGUGGAGAACUAACAGGAUAACACAUAUUCCCGUCAUCAU .....(((((((.(((.(((((((........))..))))).))).)))))))..(((......)))(((.((((.....)))))))................................. (-24.94 = -24.62 + -0.31)

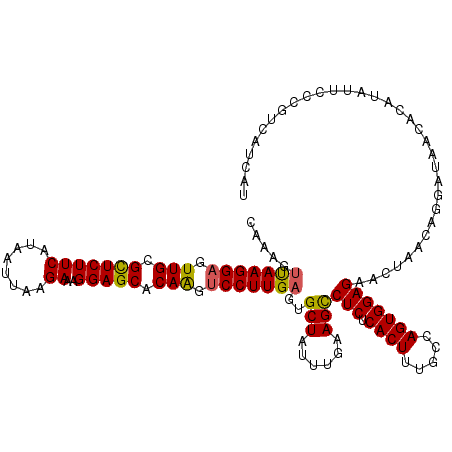
| Location | 17,488,015 – 17,488,135 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.94 |
| Mean single sequence MFE | -35.68 |
| Consensus MFE | -25.96 |
| Energy contribution | -29.65 |
| Covariance contribution | 3.69 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.73 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
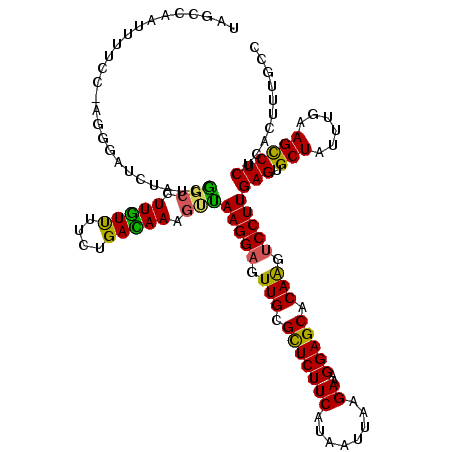
>2R_DroMel_CAF1 17488015 120 + 20766785 GGCAAAGUGAGAGACUUCAAAUAGCACUCAAGGACUUGUGCUCCUUUCUUAAUCAUGAAGAACGCAACUCCUUGACUUUAUCAGAAAACAAGGCCAAUAGAUCCCUCGGAAAAUUGGCUA .((....((((....))))....))..(((((((.(((((.((..(((........))))).))))).)))))))................(((((((...(((...)))..))))))). ( -29.10) >DroSec_CAF1 16116 103 + 1 GGCAAAGUGAGAGGCUUCAAAUAGCACUCAAGGACUUGUGCUCCUUUCUUAAUUAUGAAGAGCGCAACUCCUUAACUUUGUCAGAAAACAAGGACGAUAGAUC----------------- ((((((((..((((((......))).)))(((((.((((((((..(((........))))))))))).))))).)))))))).....................----------------- ( -35.30) >DroSim_CAF1 28433 120 + 1 GGCAAAGUGAGAGGCUUCAAAUAGCGCUCAAGGACUUGUGCUCCUUUCUCAAUUAUGAAGAGCGCAACUCCUUAACUUUGUCAGAAAACAAGGCCGAUAGAUCCCUAGGAAAAUUGGCUA ((((((((..((((((......))).)))(((((.((((((((..(((........))))))))))).))))).)))))))).........(((((((...(((...)))..))))))). ( -44.70) >DroYak_CAF1 24328 120 + 1 GGCAGAGUGAGAGGCUUCAUAUAGCACUCAAGGAUCUGUGCUCCUUUCUAAAUUAUGAAGAACGCAACGCCUUAGUUUUAUCAACAGAUAAGGCUAAUAGAUCCCUUAGAAUCCUGGCUA (.(((..((((..(((......))).)))).((((((((((...((((........))))...)).......((((((((((....)))))))))))))))))).........))).).. ( -33.60) >consensus GGCAAAGUGAGAGGCUUCAAAUAGCACUCAAGGACUUGUGCUCCUUUCUUAAUUAUGAAGAACGCAACUCCUUAACUUUAUCAGAAAACAAGGCCAAUAGAUCCCU_GGAAAAUUGGCUA ((((((((..((((((......))).)))(((((.((((((((..(((........))))))))))).))))).)))))))).........(((((((...((.....))..))))))). (-25.96 = -29.65 + 3.69)


| Location | 17,488,015 – 17,488,135 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.94 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -24.27 |
| Energy contribution | -23.28 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17488015 120 - 20766785 UAGCCAAUUUUCCGAGGGAUCUAUUGGCCUUGUUUUCUGAUAAAGUCAAGGAGUUGCGUUCUUCAUGAUUAAGAAAGGAGCACAAGUCCUUGAGUGCUAUUUGAAGUCUCUCACUUUGCC ..((((((..(((...)))...)))))).....((((.((((.(.(((((((.(((.(((((((........))..))))).))).))))))).)..)))).)))).............. ( -33.70) >DroSec_CAF1 16116 103 - 1 -----------------GAUCUAUCGUCCUUGUUUUCUGACAAAGUUAAGGAGUUGCGCUCUUCAUAAUUAAGAAAGGAGCACAAGUCCUUGAGUGCUAUUUGAAGCCUCUCACUUUGCC -----------------.......................((((((.(((((.(((.(((((((........))..))))).))).)))))(((.(((......))))))..)))))).. ( -27.60) >DroSim_CAF1 28433 120 - 1 UAGCCAAUUUUCCUAGGGAUCUAUCGGCCUUGUUUUCUGACAAAGUUAAGGAGUUGCGCUCUUCAUAAUUGAGAAAGGAGCACAAGUCCUUGAGCGCUAUUUGAAGCCUCUCACUUUGCC ..(((.((..(((...)))...)).)))............((((((.(((((.(((.((((((((....)))....))))).))).)))))(((.(((......))))))..)))))).. ( -33.90) >DroYak_CAF1 24328 120 - 1 UAGCCAGGAUUCUAAGGGAUCUAUUAGCCUUAUCUGUUGAUAAAACUAAGGCGUUGCGUUCUUCAUAAUUUAGAAAGGAGCACAGAUCCUUGAGUGCUAUAUGAAGCCUCUCACUCUGCC ....((((......((((((((....((((((...(((.....)))))))))(((.(.((((.........)))).).)))..))))))))(((.(((......))))))....)))).. ( -31.30) >consensus UAGCCAAUUUUCC_AGGGAUCUAUCGGCCUUGUUUUCUGACAAAGUUAAGGAGUUGCGCUCUUCAUAAUUAAGAAAGGAGCACAAGUCCUUGAGUGCUAUUUGAAGCCUCUCACUUUGCC .........................(((.(((((....))))).)))(((((.(((.(((((((........))..))))).))).)))))(((.(((......)))))).......... (-24.27 = -23.28 + -1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:44 2006