| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,487,007 – 17,487,124 |
| Length | 117 |
| Max. P | 0.607720 |

| Location | 17,487,007 – 17,487,124 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.87 |
| Mean single sequence MFE | -31.05 |
| Consensus MFE | -17.44 |
| Energy contribution | -20.17 |
| Covariance contribution | 2.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
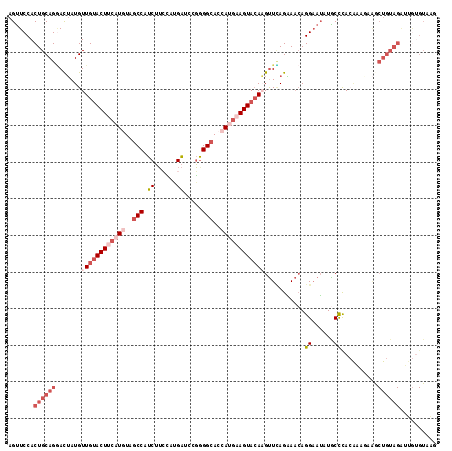
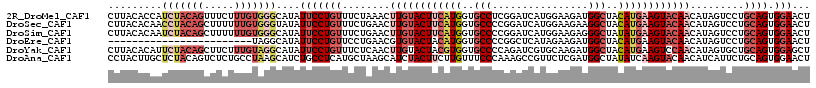
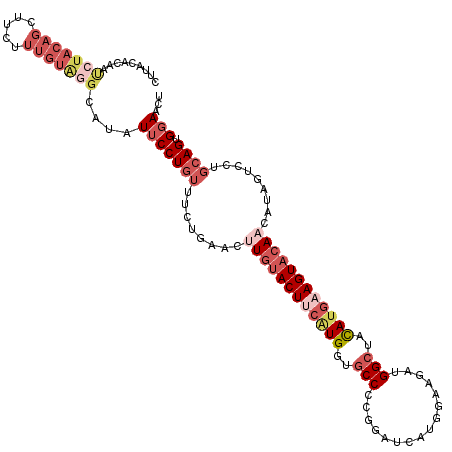
>2R_DroMel_CAF1 17487007 117 + 20766785 AGUUCCACUGCAGGACUAUGUUGUACUUCAUGUAGCCAUCUUCCAUGAUCCGAGGCACCAUGAAGUACAAGUUUAGAAACAGGAAUAUGCCCACAAAGAAACUGUAGAUGGUGUAAG ....(((((((((..(((..((((((((((((..((((((......)))....)))..))))))))))))...))).....((.......)).........)))))).)))...... ( -35.70) >DroSec_CAF1 15109 117 + 1 AGUUCCACUGCAGGACUAUGUUGUACUUCAUGUAGCCUUCUUCCAUGAUCCGGGGCACCAUGAAGUACAAGUUCAGAAACAGGAAUAUACCCACAAAAAAGCUGUAGGUUGUGUAAG .(((((....(.(((((....(((((((((((..((((((......))....))))..)))))))))))))))).).....)))))..(((.(((.......))).)))........ ( -32.20) >DroSim_CAF1 27240 117 + 1 AGUUCCACUGCAGGACUAUGUUGUACUUCAUAUAGCCCUCUUCCAUGAUCCGGGGCACCAUGAAGUACAAGUUCAGAAACAGGAAUAUGCCCACAAAAAAGCUGUAGAUUGUGUAAG .......((((((.......(((((((((((...(((((..((...))...)))))...)))))))))))((((........))))...............)))))).......... ( -30.20) >DroEre_CAF1 17708 93 + 1 AGUUCCACUGCAGGACUAUGUUGUACUUCAUGUAGCCAUCUUCUAUGAGCCGGGGCACCAUGUAGUACACGUUCAGGAACAGGAAUAUGCCUA------------------------ .(((((...(((......)))((((((.((((..(((.((......)).....)))..)))).))))))......)))))(((......))).------------------------ ( -26.70) >DroYak_CAF1 23314 117 + 1 AGCUCCACUGCAGCACUAUGUUGGACUUCAUGUAGCCAUCUUGCACGAUCUGGGGCACCACGUAGUACAAGUUGAGAAACAGGAAUAUGCCUACAAAGAAGCUGUAGAAUGUGUAAG .......(((((((.((.(((.((..(((.(((..(((.((((.((.((.(((....))).)).)).)))).)).)..))).)))....)).))).))..))))))).......... ( -30.20) >DroAna_CAF1 17867 117 + 1 AGUUCCACUGCAGAAUGAUGUUGUACUUGAUAUAGCCAUCGAGAACGGCUUUGGGAAACAAGAAGUAGAUGCUUAGCAUGAGGCAGAUGCUUAGGCAGAGACUGUAGAGCAAGUAGG .......((((((......(((((.((((((......)))))).)))))(((((....)...(((((..(((((......)))))..)))))...))))..)))))).......... ( -31.30) >consensus AGUUCCACUGCAGGACUAUGUUGUACUUCAUGUAGCCAUCUUCCAUGAUCCGGGGCACCAUGAAGUACAAGUUCAGAAACAGGAAUAUGCCCACAAAGAAGCUGUAGAUUGUGUAAG .......((((((.......((((((((((((..(((.((......)).....)))..))))))))))))...........((......))..........)))))).......... (-17.44 = -20.17 + 2.72)


| Location | 17,487,007 – 17,487,124 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.87 |
| Mean single sequence MFE | -29.67 |
| Consensus MFE | -17.59 |
| Energy contribution | -20.44 |
| Covariance contribution | 2.85 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17487007 117 - 20766785 CUUACACCAUCUACAGUUUCUUUGUGGGCAUAUUCCUGUUUCUAAACUUGUACUUCAUGGUGCCUCGGAUCAUGGAAGAUGGCUACAUGAAGUACAACAUAGUCCUGCAGUGGAACU ..............((((((.((((((((........(((....)))((((((((((((..(((((...........)).)))..))))))))))))....)))).)))).)))))) ( -35.10) >DroSec_CAF1 15109 117 - 1 CUUACACAACCUACAGCUUUUUUGUGGGUAUAUUCCUGUUUCUGAACUUGUACUUCAUGGUGCCCCGGAUCAUGGAAGAAGGCUACAUGAAGUACAACAUAGUCCUGCAGUGGAACU ........((((((((.....))))))))...(((((((..(((...((((((((((((..((((((.....))).....)))..))))))))))))..)))....)))).)))... ( -36.90) >DroSim_CAF1 27240 117 - 1 CUUACACAAUCUACAGCUUUUUUGUGGGCAUAUUCCUGUUUCUGAACUUGUACUUCAUGGUGCCCCGGAUCAUGGAAGAGGGCUAUAUGAAGUACAACAUAGUCCUGCAGUGGAACU .........(((((.((....(((((((((......)))).......((((((((((((..(((((...........).))))..)))))))))))))))))....)).)))))... ( -33.00) >DroEre_CAF1 17708 93 - 1 ------------------------UAGGCAUAUUCCUGUUCCUGAACGUGUACUACAUGGUGCCCCGGCUCAUAGAAGAUGGCUACAUGAAGUACAACAUAGUCCUGCAGUGGAACU ------------------------........(((((((..(((....((((((.((((..(((....((......))..)))..)))).))))))...)))....)))).)))... ( -22.00) >DroYak_CAF1 23314 117 - 1 CUUACACAUUCUACAGCUUCUUUGUAGGCAUAUUCCUGUUUCUCAACUUGUACUACGUGGUGCCCCAGAUCGUGCAAGAUGGCUACAUGAAGUCCAACAUAGUGCUGCAGUGGAGCU ..............((((((.((((((.(((.(((.(((...(((.(((((((.(..(((....)))..).))))))).)))..))).)))(.....)...))))))))).)))))) ( -29.90) >DroAna_CAF1 17867 117 - 1 CCUACUUGCUCUACAGUCUCUGCCUAAGCAUCUGCCUCAUGCUAAGCAUCUACUUCUUGUUUCCCAAAGCCGUUCUCGAUGGCUAUAUCAAGUACAACAUCAUUCUGCAGUGGAACU .(((((.((...........(((...(((((.......)))))..)))..(((((..(((.......(((((((...))))))))))..)))))............))))))).... ( -21.11) >consensus CUUACACAAUCUACAGCUUCUUUGUAGGCAUAUUCCUGUUUCUGAACUUGUACUUCAUGGUGCCCCGGAUCAUGGAAGAUGGCUACAUGAAGUACAACAUAGUCCUGCAGUGGAACU .........(((((((.....)))))))....(((((((........((((((((((((..(((................)))..)))))))))))).........)))).)))... (-17.59 = -20.44 + 2.85)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:40 2006