
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,859,568 – 2,859,673 |
| Length | 105 |
| Max. P | 0.707578 |

| Location | 2,859,568 – 2,859,673 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 74.56 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -13.78 |
| Energy contribution | -14.48 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2859568 105 - 20766785 UGCAAGUCAUGGGUAAGUUAAAAAUUAUACUAUUC-CCAGAGUUCC--CCAGAAUCUGGGAA-UGCUCUGGUCGAUCCUAUUUUCUUGGGUGGGUCUUAACUUUUCAGC .........((((.(((((((..............-(((((((..(--((((...)))))..-.)))))))..((.(((((((....)))))))))))))))))))).. ( -30.80) >DroVir_CAF1 11389 98 - 1 UGCAAGUCAUGGGUAAGUUAAGAAUUUUGAUUACUGCGCCAA-CUUCUCUC-AAG-----GA-AGUUGCGCGCCAGCCUAUCUUCUUGGGCGGGUU--GAAUUUUCAG- ..(((.((.(((((((.(((((...))))))))))(((((((-(((((...-..)-----))-))))).))))))(((((......))))))).))--).........- ( -29.50) >DroSec_CAF1 11253 105 - 1 UGCAAGUCAUGGGUAAGUUAAAAAUUAUACUUUUC-CCAGAGUUCC--CCAGAAUAGGGGAA-UGCUCUGGUCGAUCCUACUUUCUUGGGUGGGUCUAAACUUUUCAGC .........((((.(((((................-((((((((((--((......))))).-.)))))))..((.(((((((....)))))))))..))))))))).. ( -32.90) >DroSim_CAF1 17955 105 - 1 UGCAAGUCAUGGGUAAGUUAAAAAUUAUGCUUUUC-CCAGAGUUCC--CCAGAAUAGGGGAA-UGCUCUGGUCGAUCCUACUUUCUUGGGUGGGUCUAAACUUUUCAGC .........((((.(((((................-((((((((((--((......))))).-.)))))))..((.(((((((....)))))))))..))))))))).. ( -32.90) >DroEre_CAF1 11065 89 - 1 UGCAAGUCAUGGGUAAUUUGAAAAUGCUUAUUUUU-CCAGAG------------------AA-UGCUCUGGUCGAACCUAUUGUUUUGGGUGGGUCUAAAAUUUUCAGC .................((((((((..........-((((((------------------..-..))))))..((.((((((......))))))))....)))))))). ( -18.90) >DroYak_CAF1 13072 106 - 1 UUCAAGUUAUGGGUAAGUUGAAAAUGCUGCUUUUC-CCAGAGUUUC--CCAGUUUUGGGAAAUGGCUCUGGUCGAACCUAUUUUCUUGGGUGGGCCUCAAAUUUUCAGC ................(((((((((...(((((((-((((((....--....)))))))))).)))...((((..(((((......))))).))))....))))))))) ( -32.40) >consensus UGCAAGUCAUGGGUAAGUUAAAAAUUAUGCUUUUC_CCAGAGUUCC__CCAGAAU_GGGGAA_UGCUCUGGUCGAUCCUAUUUUCUUGGGUGGGUCUAAAAUUUUCAGC .........((((.(((((.................(((((((.....((.......)).....))))))).....(((((((....)))))))....))))))))).. (-13.78 = -14.48 + 0.70)

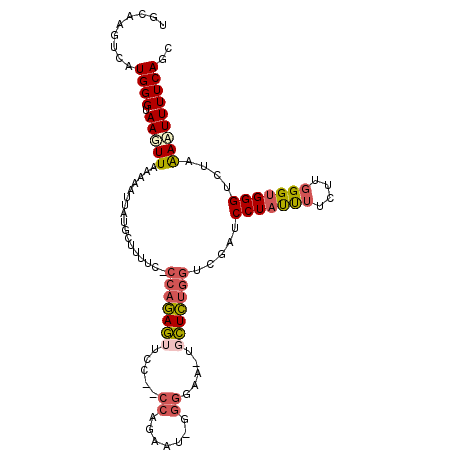
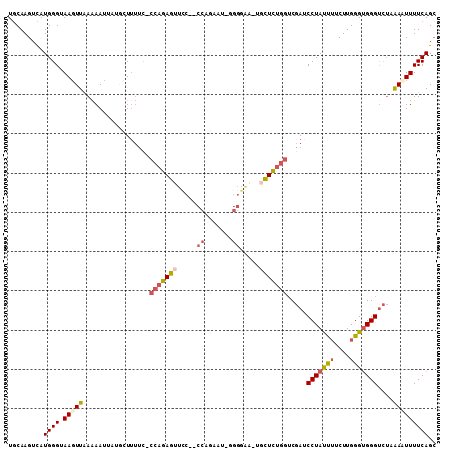
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:19 2006