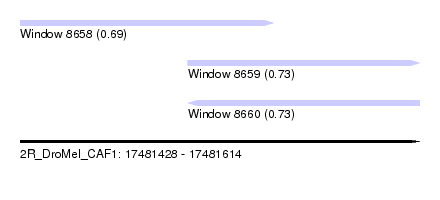
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,481,428 – 17,481,614 |
| Length | 186 |
| Max. P | 0.732698 |

| Location | 17,481,428 – 17,481,546 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -17.92 |
| Energy contribution | -19.32 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17481428 118 + 20766785 UAUCAACCUGGAGGCGGAACCAAGGUGAGUUAUGGGUUUUUCUGGGCUUACAAUUCGGAAGAUAAGAAUUGGAUUGCAAAAAAAAACCCAAUUCAUCUUCAGAGACUGCCAUUCUCUC .........((((((((....(((((((((..(((((((((.(..((...((((((.........))))))....))..).))))))))))))))))))......))))).))).... ( -35.40) >DroSec_CAF1 9550 114 + 1 UAUCAACCUGGAGGCGGAACCAAGGUGAGUUCUGGGUUUUUCUGGGUUUACAAUUCGUAAGAUAAGAGAUGGAUUGCAAAA--AAACCCAAUUCAUCUUCAGAGACUGCC--UCUCAC .........((((((((....(((((((((..(((((((((.((......((((((((..........)))))))))).))--))))))))))))))))......)))))--)))... ( -35.30) >DroSim_CAF1 21664 115 + 1 UAUCAACCUGGAGGCGGAACCAAGGUGAGUUCUGGGUUUUUCUGGGUUUACAAUUCGUAAGAUAAGAGAUGGAGUGCAAAA-AAAACCCAUUUCAUCUAUAGAGACUGCC--UCUCAC .........((((((((((((.....).))))).(((((((.(((((.......((....))...(((((((.((......-...)))))))))))))).))))))))))--)))... ( -33.00) >DroEre_CAF1 12066 115 + 1 UCUCAACCUGGAGGCGGAACCAAGGUGAGUUCUUGUUUUUCCCUGAAAUACAAUUCAGAAGAUAAGAGCUGGCUUCGGGA---AUACUCAAUUCAUUCUCAGAGACUGCCUUUCUGUC .....((..((((((((..((.((((.((((((((((((...(((((......))))))))))))))))).)))).))((---((.....))))...........))))))))..)). ( -36.50) >DroYak_CAF1 17648 107 + 1 UCUCAAUCUGGAGGCGGAGCCACGGUGAGUUCUUGGUUUUUC--------CAAUUCGGAAGAUAAGAGUUGGAUUGCAAA---AUUCCCCAUUCAUCUUCAGAGAAUGCCUAUCUGUC ......(((((((..(((.....((.(((((....((...((--------((((((.........))))))))..))..)---))))))..)))..)))))))............... ( -26.80) >consensus UAUCAACCUGGAGGCGGAACCAAGGUGAGUUCUGGGUUUUUCUGGGUUUACAAUUCGGAAGAUAAGAGAUGGAUUGCAAAA__AAACCCAAUUCAUCUUCAGAGACUGCC__UCUCUC .........((((((((((((.(((.....))).))))...............(((.((((((..(((((((...............))))))))))))).))).)))))..)))... (-17.92 = -19.32 + 1.40)

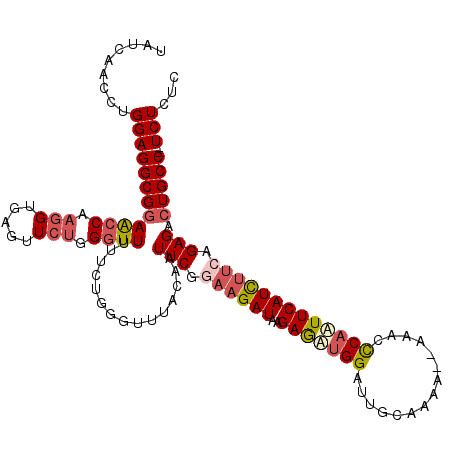
| Location | 17,481,506 – 17,481,614 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.02 |
| Mean single sequence MFE | -22.04 |
| Consensus MFE | -13.79 |
| Energy contribution | -14.75 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17481506 108 + 20766785 AAAAAAAACCCAAUUCAUCUUCAGAGACUGCCAUUCUCUCUAUAU--------AUUGGCCUUUUGGUAUGGAGCUGGCUCCAUGCAGAUAAAA----UGAAACCUUAUAUCUUUGUUUGU .....................((((((..((((............--------..))))......((((((((....))))))))........----............))))))..... ( -21.74) >DroSec_CAF1 9628 104 + 1 AAA--AAACCCAAUUCAUCUUCAGAGACUGCC--UCUCACUAUGU--------AUGGGCCUUUUGGUAUGGAGCUUGCUGCAUACAGAUAAAA----UGAAACCCUAUAUCUUUGUUUGU (((--(..((((...(((....((((.....)--)))....))).--------.))))..)))).(((((.((....)).)))))(((((...----..........)))))........ ( -18.22) >DroSim_CAF1 21742 105 + 1 AAA-AAAACCCAUUUCAUCUAUAGAGACUGCC--UCUCACUAUGU--------AUGGGCCUUUUGGUAUGGAGCUUGCUCCAUACAGAUAAAA----UGAAACCCUAUAUCUUUGUUUGU .((-((..(((((..(((....((((.....)--)))....))).--------)))))..)))).((((((((....))))))))(((((...----..........)))))........ ( -24.62) >DroEre_CAF1 12144 108 + 1 GA---AUACUCAAUUCAUUCUCAGAGACUGCCUUUCUGUCUAUAU--------AUAGGCCUU-UCCAAUGGAGCUUGCUCCAUACUGAUAGAAUGAAUGAAACCCCAUAUCUUUGUUUAU ..---....(((.(((((((((((.....((((............--------..))))...-....((((((....)))))).))))..)))))))))).................... ( -24.14) >DroYak_CAF1 17718 113 + 1 AA---AUUCCCCAUUCAUCUUCAGAGAAUGCCUAUCUGUCUAUAUAUGUACAUAUAGACCUCUUAGUAUGAAGCUUGCUCCAUACUGAUAACA----UGAAACCCUAUAUCUUUGUUUAU ..---................((((((..........((((((((......))))))))...((((((((.((....)).)))))))).....----............))))))..... ( -21.50) >consensus AAA__AAACCCAAUUCAUCUUCAGAGACUGCC__UCUCUCUAUAU________AUAGGCCUUUUGGUAUGGAGCUUGCUCCAUACAGAUAAAA____UGAAACCCUAUAUCUUUGUUUGU .....................((((((..((((......................))))......((((((((....))))))))........................))))))..... (-13.79 = -14.75 + 0.96)

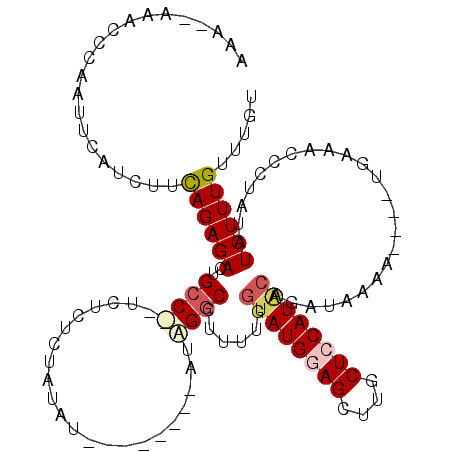
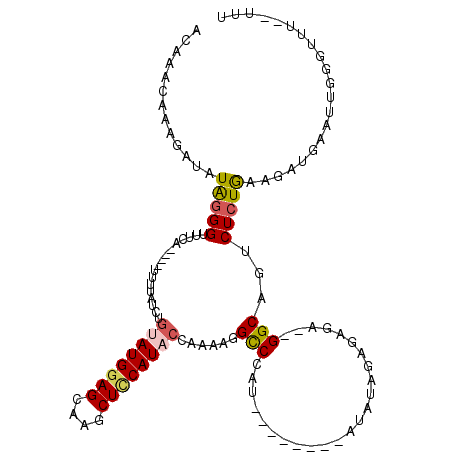
| Location | 17,481,506 – 17,481,614 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.02 |
| Mean single sequence MFE | -26.94 |
| Consensus MFE | -12.70 |
| Energy contribution | -13.10 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17481506 108 - 20766785 ACAAACAAAGAUAUAAGGUUUCA----UUUUAUCUGCAUGGAGCCAGCUCCAUACCAAAAGGCCAAU--------AUAUAGAGAGAAUGGCAGUCUCUGAAGAUGAAUUGGGUUUUUUUU ........(((((.((.......----)).)))))..((((((....))))))...(((((.(((((--------.(((..(((((.......)))))....))).))))).)))))... ( -22.40) >DroSec_CAF1 9628 104 - 1 ACAAACAAAGAUAUAGGGUUUCA----UUUUAUCUGUAUGCAGCAAGCUCCAUACCAAAAGGCCCAU--------ACAUAGUGAGA--GGCAGUCUCUGAAGAUGAAUUGGGUUU--UUU ........(((((..........----...)))))(((((.((....)).))))).((((((((((.--------.(((..(.(((--(.....)))).)..)))...)))))))--))) ( -24.82) >DroSim_CAF1 21742 105 - 1 ACAAACAAAGAUAUAGGGUUUCA----UUUUAUCUGUAUGGAGCAAGCUCCAUACCAAAAGGCCCAU--------ACAUAGUGAGA--GGCAGUCUCUAUAGAUGAAAUGGGUUUU-UUU ........(((((..........----...)))))((((((((....)))))))).(((((((((((--------.(((....(((--(.....))))....)))..)))))))))-)). ( -30.52) >DroEre_CAF1 12144 108 - 1 AUAAACAAAGAUAUGGGGUUUCAUUCAUUCUAUCAGUAUGGAGCAAGCUCCAUUGGA-AAGGCCUAU--------AUAUAGACAGAAAGGCAGUCUCUGAGAAUGAAUUGAGUAU---UC ...............(((((((((((((((..((((.((((((....)))))).(((-...((((..--------............))))..)))))))))))))).)))).))---)) ( -26.04) >DroYak_CAF1 17718 113 - 1 AUAAACAAAGAUAUAGGGUUUCA----UGUUAUCAGUAUGGAGCAAGCUUCAUACUAAGAGGUCUAUAUGUACAUAUAUAGACAGAUAGGCAUUCUCUGAAGAUGAAUGGGGAAU---UU ...............(((((((.----..((((((((((((((....))))))))).....(((((((((....))))))))).))))).(((((((....)).))))).)))))---)) ( -30.90) >consensus ACAAACAAAGAUAUAGGGUUUCA____UUUUAUCUGUAUGGAGCAAGCUCCAUACCAAAAGGCCCAU________AUAUAGAGAGA__GGCAGUCUCUGAAGAUGAAUUGGGUUU__UUU .............(((((.................((((((((....))))))))......(((........................)))...)))))..................... (-12.70 = -13.10 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:35 2006