| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,477,258 – 17,477,385 |
| Length | 127 |
| Max. P | 0.713916 |

| Location | 17,477,258 – 17,477,353 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 98.60 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -25.97 |
| Energy contribution | -25.83 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17477258 95 + 20766785 ACUUUAACUGUAAAAAUGUUUAUGGCGAACAGAAGCGCCAUCUAGCGCUCGUUAAGUGGCAACUGCAGACGCCGCUGCGUCGCGCAUUUCAAGUG ......(((....((((((..((((((........))))))...(((..(((..((((((..(....)..))))))))).)))))))))..))). ( -27.80) >DroSec_CAF1 5532 95 + 1 ACUUUAACUGUAAAAAUGUUUAUGGCGAACAGAAGCGCCAUCUAGCGCUCGUUAAGUGGCAACUGCAGACGCCGCUGCGUCGCGCAUUUCAAGUG ......(((....((((((..((((((........))))))...(((..(((..((((((..(....)..))))))))).)))))))))..))). ( -27.80) >DroSim_CAF1 14099 95 + 1 ACUUUAACUGUAAAAAUGUUUAUGGCGAACAGAAGCGCCAUCUAGCGCUCGUUAAGUGGCAACUGCAGACGCCGCUGCGUCGCGCAUUUCAAGUG ......(((....((((((..((((((........))))))...(((..(((..((((((..(....)..))))))))).)))))))))..))). ( -27.80) >DroEre_CAF1 5497 95 + 1 ACUUUAACUGUAAAAAUGUUUAUGGCGAACAGAAGCGCCAUCUAGCGCUCGUUAAGUGGCAACUGCAGACGCCGCUGCGUCGCGCAUUUCAAGUG ......(((....((((((..((((((........))))))...(((..(((..((((((..(....)..))))))))).)))))))))..))). ( -27.80) >DroYak_CAF1 13447 95 + 1 ACUUUAACUGUAAAAAUGUUUAUGGCGAACAGAAGCGCCAUCUAGCGCUCGUUAAGUGGCAACUGCAGACGCCGCUGCGUCGCGCAUUUCAAGUG ......(((....((((((..((((((........))))))...(((..(((..((((((..(....)..))))))))).)))))))))..))). ( -27.80) >DroAna_CAF1 5140 95 + 1 ACUUUAACUGUAAAAAGGUUUAUGGCAAACAGAAGCGCCAUCUAGUGCUCGUUAAGUGGCAACUGCAGACGCCGUUGCGUCGCGCAUUUCAAGUG ((((.((.(((....((((((.((.....)).))))((((..(((......)))..))))..))((.(((((....)))))))))).)).)))). ( -22.90) >consensus ACUUUAACUGUAAAAAUGUUUAUGGCGAACAGAAGCGCCAUCUAGCGCUCGUUAAGUGGCAACUGCAGACGCCGCUGCGUCGCGCAUUUCAAGUG ((((.((.(((......((((.((.....)).))))((((..((((....))))..))))....((.(((((....)))))))))).)).)))). (-25.97 = -25.83 + -0.14)

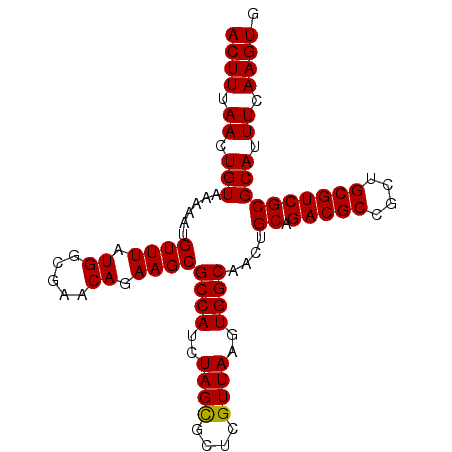
| Location | 17,477,281 – 17,477,385 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 96.60 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -29.85 |
| Energy contribution | -29.74 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17477281 104 + 20766785 GGCGAACAGAAGCGCCAUCUAGCGCUCGUUAAGUGGCAACUGCAGACGCCGCUGCGUCGCGCAUUUCAAGUGGAAAAAUGCCCGGAAAACUGGGGAAAAUCCGC ((((........)))).....((((.(((..((((((..(....)..)))))))))..)))).......(((((....(.(((((....))))).)...))))) ( -37.50) >DroSec_CAF1 5555 104 + 1 GGCGAACAGAAGCGCCAUCUAGCGCUCGUUAAGUGGCAACUGCAGACGCCGCUGCGUCGCGCAUUUCAAGUGGAAAAAUACCCGGAAAACUGGAGAAAAUCCGC ((((........)))).....((((.(((..((((((..(....)..)))))))))..)))).......(((((.......((((....))))......))))) ( -33.32) >DroSim_CAF1 14122 104 + 1 GGCGAACAGAAGCGCCAUCUAGCGCUCGUUAAGUGGCAACUGCAGACGCCGCUGCGUCGCGCAUUUCAAGUGGAAAAAUGCCCGGAAAACUGGAGAAAAUCCGC ((((........)))).....((((.(((..((((((..(....)..)))))))))..)))).......(((((......(((((....)))).)....))))) ( -33.70) >DroEre_CAF1 5520 104 + 1 GGCGAACAGAAGCGCCAUCUAGCGCUCGUUAAGUGGCAACUGCAGACGCCGCUGCGUCGCGCAUUUCAAGUGGAAAAAUGCCCGGAAAACUGAGCAAAAUCCGC ((((........)))).....((((.(((..((((((..(....)..)))))))))..)))).......(((((....(((.(((....))).)))...))))) ( -35.10) >DroYak_CAF1 13470 104 + 1 GGCGAACAGAAGCGCCAUCUAGCGCUCGUUAAGUGGCAACUGCAGACGCCGCUGCGUCGCGCAUUUCAAGUGGAAAAAUGCCCGGAAAACUGAGGAAAAUCCGC ((((........)))).....((((.(((..((((((..(....)..)))))))))..)))).......(((((......(((((....))).))....))))) ( -33.70) >DroAna_CAF1 5163 104 + 1 GGCAAACAGAAGCGCCAUCUAGUGCUCGUUAAGUGGCAACUGCAGACGCCGUUGCGUCGCGCAUUUCAAGUGGAAAACUGCCCGGAAAACUGUGAAAAAUCCGC ((((.......((((((..(((......)))..))))....((.(((((....))))))))).((((.....))))..))))((((.............)))). ( -27.82) >consensus GGCGAACAGAAGCGCCAUCUAGCGCUCGUUAAGUGGCAACUGCAGACGCCGCUGCGUCGCGCAUUUCAAGUGGAAAAAUGCCCGGAAAACUGGGGAAAAUCCGC ((((.......((((((..((((....))))..))))....((.(((((....))))))))).((((.....))))..))))((((.............)))). (-29.85 = -29.74 + -0.11)

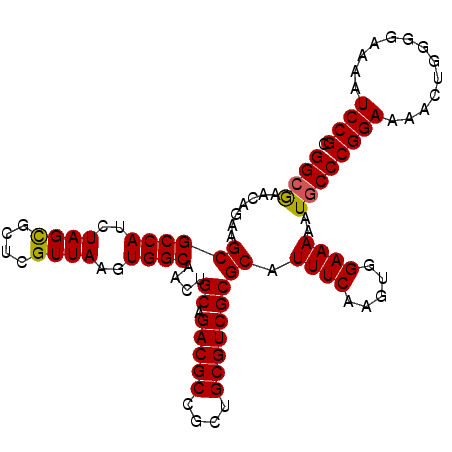

| Location | 17,477,281 – 17,477,385 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 96.60 |
| Mean single sequence MFE | -33.43 |
| Consensus MFE | -33.41 |
| Energy contribution | -33.30 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.60 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17477281 104 - 20766785 GCGGAUUUUCCCCAGUUUUCCGGGCAUUUUUCCACUUGAAAUGCGCGACGCAGCGGCGUCUGCAGUUGCCACUUAACGAGCGCUAGAUGGCGCUUCUGUUCGCC ((((((........(((....(((((((((.......)))))))(((((((....))))).)).....))....)))(((((((....)))))))..)))))). ( -34.90) >DroSec_CAF1 5555 104 - 1 GCGGAUUUUCUCCAGUUUUCCGGGUAUUUUUCCACUUGAAAUGCGCGACGCAGCGGCGUCUGCAGUUGCCACUUAACGAGCGCUAGAUGGCGCUUCUGUUCGCC ((((((........(((....(((((((((.......)))))))(((((((....))))).)).....))....)))(((((((....)))))))..)))))). ( -32.90) >DroSim_CAF1 14122 104 - 1 GCGGAUUUUCUCCAGUUUUCCGGGCAUUUUUCCACUUGAAAUGCGCGACGCAGCGGCGUCUGCAGUUGCCACUUAACGAGCGCUAGAUGGCGCUUCUGUUCGCC ((((((........(((....(((((((((.......)))))))(((((((....))))).)).....))....)))(((((((....)))))))..)))))). ( -34.90) >DroEre_CAF1 5520 104 - 1 GCGGAUUUUGCUCAGUUUUCCGGGCAUUUUUCCACUUGAAAUGCGCGACGCAGCGGCGUCUGCAGUUGCCACUUAACGAGCGCUAGAUGGCGCUUCUGUUCGCC ((((((........(((....(((((((((.......)))))))(((((((....))))).)).....))....)))(((((((....)))))))..)))))). ( -34.90) >DroYak_CAF1 13470 104 - 1 GCGGAUUUUCCUCAGUUUUCCGGGCAUUUUUCCACUUGAAAUGCGCGACGCAGCGGCGUCUGCAGUUGCCACUUAACGAGCGCUAGAUGGCGCUUCUGUUCGCC ((((((........(((....(((((((((.......)))))))(((((((....))))).)).....))....)))(((((((....)))))))..)))))). ( -34.90) >DroAna_CAF1 5163 104 - 1 GCGGAUUUUUCACAGUUUUCCGGGCAGUUUUCCACUUGAAAUGCGCGACGCAACGGCGUCUGCAGUUGCCACUUAACGAGCACUAGAUGGCGCUUCUGUUUGCC ((((((........(((.....(((((.((((.....))))((((.(((((....))))))))).)))))....)))((((.((....)).))))..)))))). ( -28.10) >consensus GCGGAUUUUCCCCAGUUUUCCGGGCAUUUUUCCACUUGAAAUGCGCGACGCAGCGGCGUCUGCAGUUGCCACUUAACGAGCGCUAGAUGGCGCUUCUGUUCGCC ((((((........(((.....((((..((((.....))))((((.(((((....)))))))))..))))....)))(((((((....)))))))..)))))). (-33.41 = -33.30 + -0.11)

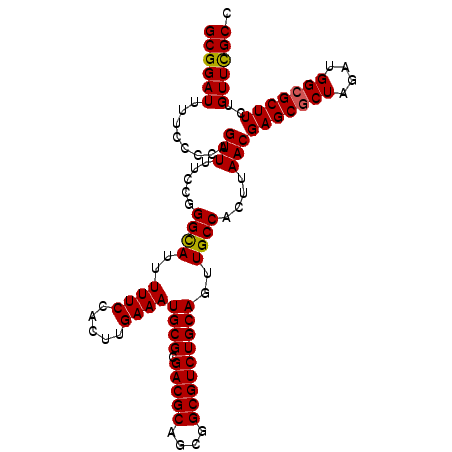
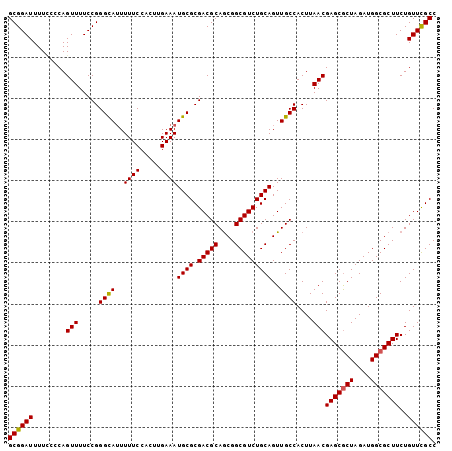
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:31 2006