
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,476,824 – 17,476,941 |
| Length | 117 |
| Max. P | 0.559463 |

| Location | 17,476,824 – 17,476,941 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.14 |
| Mean single sequence MFE | -28.63 |
| Consensus MFE | -16.88 |
| Energy contribution | -17.72 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17476824 117 + 20766785 AAGUCAAGGUUGAAAUAGACAACCUGUUGCGUAAUUGAAAAGCGGGAAGUAAUUUGCUGGG-GAGACCAGAUUACGGACAGGUUGACGUUGAUAAGCUGGAUAAAUGUUUUGAACAAA .......(.(..(((((..(((((((((.(((((((....((((((......)))))).((-....)).))))))))))))))))..(((....)))........)))))..).)... ( -32.50) >DroSec_CAF1 5070 118 + 1 AAGUCAAGGUUGAAAUAGACAACCUGUUGCGUAAUUGAAAAGCUGGAAGUAAUUUGCUGGGCGAGACCAGAUUACGGACAGGUUGACGUUGAUAAGCUGGAUAAAUGUUUUAAACAAA .......(.((((((((..(((((((((.(((((((.....(((...(((.....))).))).......))))))))))))))))..(((....)))........)))))))).)... ( -29.10) >DroSim_CAF1 13638 114 + 1 AAGUCAA-GUUGAAAUAGACAACCUGUUGCGUAAUUGAAAAGCUGGAAGUAAUUUGCUGGGCGAGACCAGAUUACG-ACAGGUUGACGUUGA-AAGCUGGAUA-AUGUUUUGAACAAA ..(((..-(((......)))(((((((..(((((((.....(((...(((.....))).))).......)))))))-))))))))))(((.(-((((......-..))))).)))... ( -30.20) >DroEre_CAF1 5025 118 + 1 AAGUCAAGGUUGAAAUAGACAACCUGUUGCGUAAUUGAAUAGGUGGAAGUAAUUUGCUGGGGGAGACAAGAUUACAGACAGGUUGACGUUGAUAAGCUGGAUAAAUGUUUUGAACAAA .......(.(..(((((..(((((((((..((((((.....((..((.....))..))...(....)..)))))).)))))))))..(((....)))........)))))..).)... ( -27.40) >DroYak_CAF1 12949 118 + 1 AAGUCAAGGUUGAAAUAGACAACCUGUUGCGUAAUUGAAAAGGUGGAAGUCAUUUGCUGGCGGAGACAAGAUUACAGACAGGUUGACGUUGAUAAGCGGGAUAAAUGUUUUGAACAAA .......(.(..(((((..(((((((((..((((((............((((.....))))(....)..)))))).))))))))).((((....)))).......)))))..).)... ( -30.60) >DroAna_CAF1 4802 97 + 1 AAGAGG----AGAAAU-GACAACCUGUUGAAUAUUUGACG-------AGUAAUUUG------GA---AAUAUUUCGGACAGGUUGACUUUGACGAGAUGCUUAAAUAUUUCCGAUAGC ....((----(((...-..(((((((((((((((((..((-------((...))))------.)---)))))))..))))))))).(((....)))...........)))))...... ( -22.00) >consensus AAGUCAAGGUUGAAAUAGACAACCUGUUGCGUAAUUGAAAAGCUGGAAGUAAUUUGCUGGG_GAGACAAGAUUACGGACAGGUUGACGUUGAUAAGCUGGAUAAAUGUUUUGAACAAA .........((((((((..((((((((...((((((...........(((.....)))...........))))))..))))))))..(((....)))........))))))))..... (-16.88 = -17.72 + 0.84)

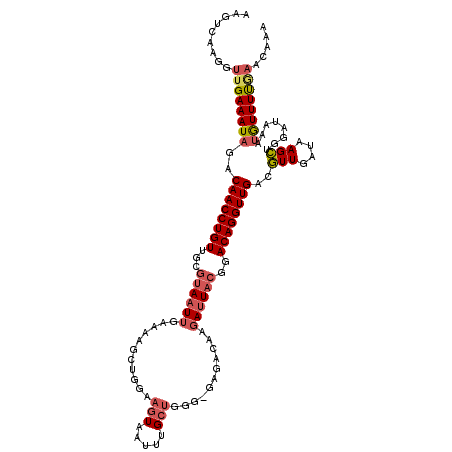
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:28 2006