
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,475,302 – 17,475,515 |
| Length | 213 |
| Max. P | 0.706399 |

| Location | 17,475,302 – 17,475,414 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -36.42 |
| Consensus MFE | -23.07 |
| Energy contribution | -22.97 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.706399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
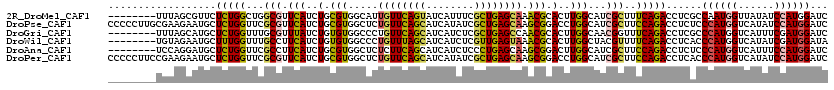
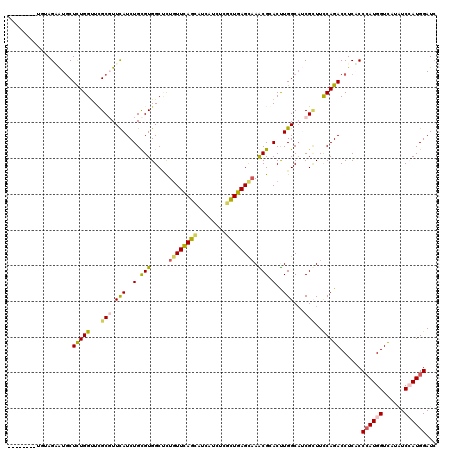
>2R_DroMel_CAF1 17475302 112 + 20766785 --------UUUAGCGUUCUCUGGCUGGCGUUCAUCUGCGUGGCAUUGUUCAGUAUCAUUUCGCUGAGCAAACGCACUUGGCAUCGCUUUCAGACCUCGCCAAUGGUUAUAUCCAUGGAUC --------....(((...(((((..((((.(((..(((((....(((((((((........))))))))))))))..)))...)))).)))))...)))..((((......))))..... ( -36.00) >DroPse_CAF1 4898 120 + 1 CCCCCUUGCGAAGAAUGCUCUGGUUCGCGUUCAUCUGCGUGGCUCUGUUCAGCAUCAUAUCGCUGAGCAAGCGGACCUGGCAUCGCUUCCAGACCUCUCCCAUGGUCAUAUCCAUGGAUC .....(((.((((.(((((..(((((((((......)))).(((.((((((((........)))))))))))))))).)))))..))))))).......((((((......))))))... ( -46.00) >DroGri_CAF1 7292 112 + 1 --------UUUAGCAUGCUCUGGUUUGCGUUUAUCUGUGUGGCCCUGUUCAGCAUCAUCUCGCUGAGCCAACGCACUUGGCAACGGUUUCAGACCUCGCCCAUGGUCAUUUCGAUGGAUC --------....(((.((....)).)))(((((((...((((((..(((((((........)))))))..........(((...((((...))))..)))...))))))...))))))). ( -32.60) >DroWil_CAF1 9590 112 + 1 --------UGUAGAAUGCUUUGGUUUGCCUUCAUCUGUGUGGCCCUGUUUAGCAUCAUCUCGUUGAGUAAACGCACUUGGCUACGUUUUCAGACCUCACCCAUGGUCAUAUCGAUGGAUA --------....(((.((........)).)))((((((((((((.((((((...(((......))).)))))).....)))))).......((((........))))......)))))). ( -26.20) >DroAna_CAF1 3357 112 + 1 --------UCCAGGAUGCUCUGGUUCGCCUUCAUCUGCGUGGCUCUCUUCAGCAUCAUCUCCCUGAGCAAGCGGACUUGGCAUCGCUUCCAGACCUCUCCCAUGGUCAUUUCCAUGGAUC --------((((.((((((..(((((((...(((....)))((((...................))))..))))))).)))))).......((((........)))).......)))).. ( -33.21) >DroPer_CAF1 4910 120 + 1 CCCCCUUCCGAAGAAUGCUCUGGUUCGCGUUCAUCUGCGUGGCUCUGUUCAGCAUCAUAUCGCUGAGCAAGCGGACCUGGCAUCGCUUCCAGACCUCACCCAUGGUCAUAUCCAUGGAUC .........((((.(((((..(((((((((......)))).(((.((((((((........)))))))))))))))).)))))..))))..........((((((......))))))... ( -44.50) >consensus ________UGUAGAAUGCUCUGGUUCGCGUUCAUCUGCGUGGCUCUGUUCAGCAUCAUCUCGCUGAGCAAACGCACUUGGCAUCGCUUCCAGACCUCACCCAUGGUCAUAUCCAUGGAUC ..................(((((...(((.(((..(.(((.....((((((((........)))))))).))).)..)))...)))..)))))......((((((......))))))... (-23.07 = -22.97 + -0.11)

| Location | 17,475,414 – 17,475,515 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.96 |
| Mean single sequence MFE | -33.83 |
| Consensus MFE | -20.55 |
| Energy contribution | -20.67 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
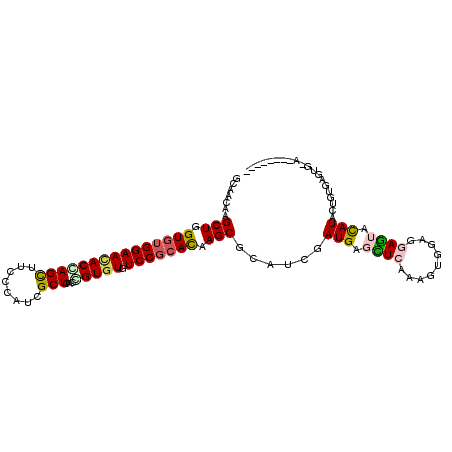
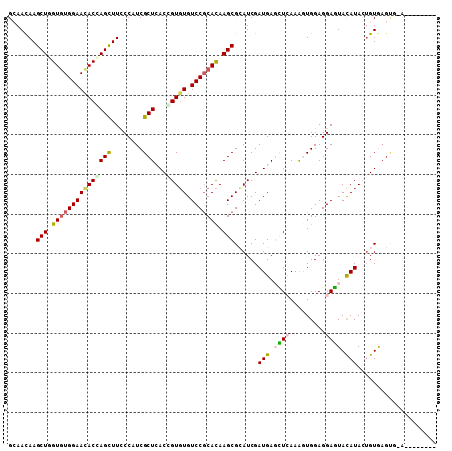
>2R_DroMel_CAF1 17475414 101 + 20766785 GGAAUAAGCUGGUCUGGAACACCAGCUUUCCAUCGCUCACAGUCUGUCCGCAUAAGCGGAUCGAUGAGUUGAAAGUGGAGGAGUACAUACUGUGAGUUAAA------- ((((..(((((((.......)))))))))))...((((((((((.((((((....)))))).))..........(((........))).))))))))....------- ( -37.00) >DroPse_CAF1 5018 107 + 1 GCAACAAGCUGGUGUGGAACACGAGCUUCCCCUCGCUGACCGUGUGUCCGCACAAGCGCAUCGAUGAGCUCAAAGUGGAGGAGUACAUACUGUGAGUG-AGUCCCAGA .......(((.(((((((((((((((........)))...))))).))))))).))).....(((..(((((.((((..(.....).)))).))))).-.)))..... ( -35.70) >DroGri_CAF1 7404 99 + 1 GCAAUAAGCUGGUGUGGAAUACUAGUUUUCCAUCGCUCACGGUGUGUCCCCAUAAGCGCAUCGAUGAUAUCAAACUGGAGGAUUAUAUACUGUGAGUG-U-------- .....((((((((((...)))))))))).....(((((((((((((((((((.......(((...))).......))).))))...))))))))))))-.-------- ( -30.24) >DroEre_CAF1 3645 94 + 1 GGAACAAGCUGGUGUGGAACACCAGCUUUCCCUCGCUCACGGUGUGUCCGCACAAGCGGAUCGAUGAGCUGAAGGUGGAGGAGUACAUAGUGUG-------------- ...(((((((((((.....))))))))((((..((((..((((.(((((((....))))).))....))))..))))..)))).......))).-------------- ( -37.30) >DroMoj_CAF1 8850 99 + 1 GCAAUAAGCUGGUGUGGAACACCAGCUUCCCAUCGCUCACCGUGUGUCCGCACAAGCGCAUCGAUGACAUCAAGCUGGAGGAUUACAUAGAGUAAGCC-U-------- ((...(((((((((.....)))))))))..(((((.......((((....)))).......))))).......))...(((.((((.....)))).))-)-------- ( -30.64) >DroPer_CAF1 5030 107 + 1 GCAACAAGCUGGUGUGGAACACGAGCUUCCCCUCGCUGACCGUGUGUCCGCACAAGCGCAUCGAUGAGCUCAAAGUGGAGGAGUACAUACUGUAAGUG-AGUCCCAGA .....(((((.(((.....))).)))))...((((((.((.((((((((.(((.(((.((....)).)))....)))..)))...))))).)).))))-))....... ( -32.10) >consensus GCAACAAGCUGGUGUGGAACACCAGCUUCCCAUCGCUCACCGUGUGUCCGCACAAGCGCAUCGAUGAGCUCAAAGUGGAGGAGUACAUACUGUGAGUG_A________ .......(((.(((((((((((((((........)))...))))).))))))).)))......(((.((((.........)))).))).................... (-20.55 = -20.67 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:27 2006