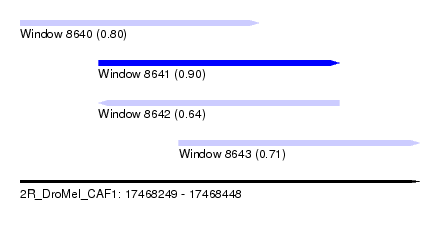
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,468,249 – 17,468,448 |
| Length | 199 |
| Max. P | 0.902682 |

| Location | 17,468,249 – 17,468,368 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.21 |
| Mean single sequence MFE | -38.51 |
| Consensus MFE | -34.19 |
| Energy contribution | -34.30 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17468249 119 + 20766785 GUUUCCGCAACAUUUUCGCAUUGGCAGCG-GGAAAGUGCAUUUAAUUAAAUUUCCCUUUGUUCGAUGCGAGUGCAAAAAGCAUUGCCUACUUUUGCGGGCGGGCAAGAAACUGUUGCACA ......((((((.((((((((((((((.(-(((((...............)))))).))).))))))).(((((.....)))))(((..((......))..)))..)))).))))))... ( -41.46) >DroSim_CAF1 4194 119 + 1 GUUUCCGCAACAUUUUCGCAGUGGCAGCG-GGAAAGUGCAUUUAAUUAAAUUUCCCUUUGUUCGAUGCGAGUGCAAAAAGCAUUGCCUACUUUUGCGGGCGGGCAAGAAACUGUUGCACA ......((((((.(((((((..(((((.(-(((((...............)))))).)))))...))).(((((.....)))))(((..((......))..)))..)))).))))))... ( -37.86) >DroYak_CAF1 4461 120 + 1 GUAUCCGCAACAUUUUCGCAGUGGCAGCAGAGAAAGUGCAUUUAAUUAAAUUUCCCUUUGUUCGAUGCGAGUGCAAAAAGCAUUGCCUACUUUUGCGGGCGGGCAAGAAACUGUUGCACA ......((((((.(((((((.(.(((((((((.((((............))))..))))))....))).).))).....((.(((((..(....)..)))))))..)))).))))))... ( -36.20) >consensus GUUUCCGCAACAUUUUCGCAGUGGCAGCG_GGAAAGUGCAUUUAAUUAAAUUUCCCUUUGUUCGAUGCGAGUGCAAAAAGCAUUGCCUACUUUUGCGGGCGGGCAAGAAACUGUUGCACA ......((((((.(((((((.(.(((.((.(.((((...((((....))))....)))).).)).))).).))).....((.(((((..(....)..)))))))..)))).))))))... (-34.19 = -34.30 + 0.11)

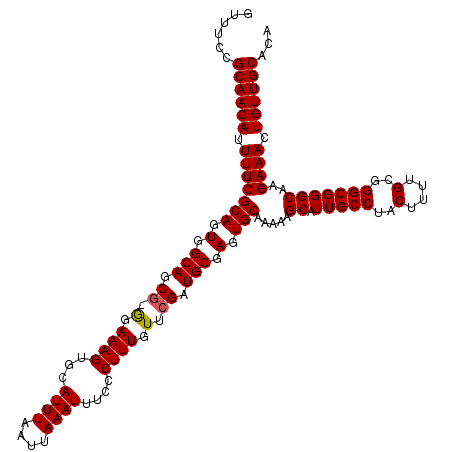
| Location | 17,468,288 – 17,468,408 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -35.27 |
| Consensus MFE | -35.49 |
| Energy contribution | -35.27 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.96 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17468288 120 + 20766785 UUUAAUUAAAUUUCCCUUUGUUCGAUGCGAGUGCAAAAAGCAUUGCCUACUUUUGCGGGCGGGCAAGAAACUGUUGCACAUGCAGCCGCAAAAUGUCGGCUGCUCAUGACAACACCAACC ................(((((((......(((((.....)))))(((..(....)..))))))))))....(((((((...((((((((.....).)))))))...)).)))))...... ( -35.90) >DroSim_CAF1 4233 120 + 1 UUUAAUUAAAUUUCCCUUUGUUCGAUGCGAGUGCAAAAAGCAUUGCCUACUUUUGCGGGCGGGCAAGAAACUGUUGCACAUGCAGCCGCAAAAUGUCGGCUGCUCAUGACAACACCAACC ................(((((((......(((((.....)))))(((..(....)..))))))))))....(((((((...((((((((.....).)))))))...)).)))))...... ( -35.90) >DroYak_CAF1 4501 120 + 1 UUUAAUUAAAUUUCCCUUUGUUCGAUGCGAGUGCAAAAAGCAUUGCCUACUUUUGCGGGCGGGCAAGAAACUGUUGCACAUGCAGCCGCAAAAUGUUGGCUGCUCAUGACAACACCAACC ................(((((((......(((((.....)))))(((..(....)..))))))))))....(((((((...((((((((.....).)))))))...)).)))))...... ( -34.00) >consensus UUUAAUUAAAUUUCCCUUUGUUCGAUGCGAGUGCAAAAAGCAUUGCCUACUUUUGCGGGCGGGCAAGAAACUGUUGCACAUGCAGCCGCAAAAUGUCGGCUGCUCAUGACAACACCAACC ................(((((((......(((((.....)))))(((..(....)..))))))))))....(((((((...((((((((.....).)))))))...)).)))))...... (-35.49 = -35.27 + -0.22)

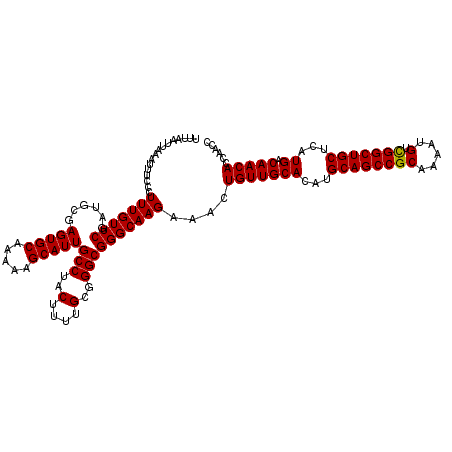
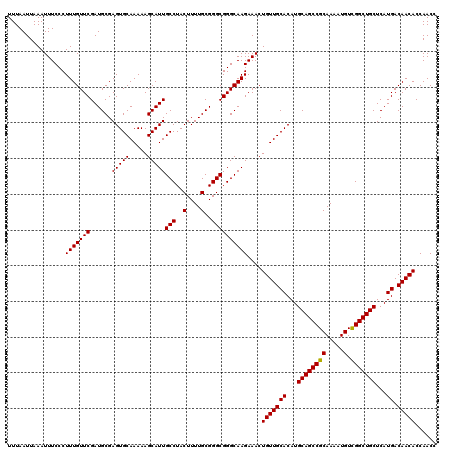
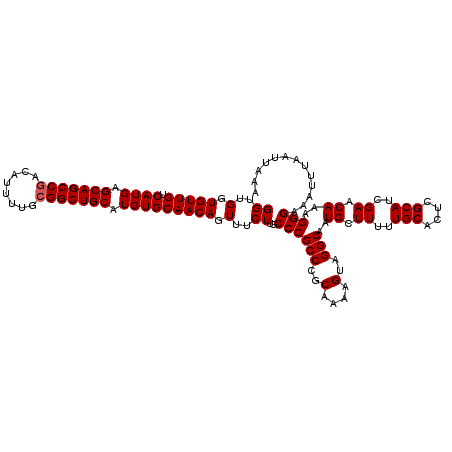
| Location | 17,468,288 – 17,468,408 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -36.53 |
| Consensus MFE | -35.93 |
| Energy contribution | -36.27 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17468288 120 - 20766785 GGUUGGUGUUGUCAUGAGCAGCCGACAUUUUGCGGCUGCAUGUGCAACAGUUUCUUGCCCGCCCGCAAAAGUAGGCAAUGCUUUUUGCACUCGCAUCGAACAAAGGGAAAUUUAAUUAAA ((..(.(((((.((((.(((((((.(.....)))))))).))))))))).)..))..((((((..(....)..)))..((.((..(((....)))..)).))..)))............. ( -37.60) >DroSim_CAF1 4233 120 - 1 GGUUGGUGUUGUCAUGAGCAGCCGACAUUUUGCGGCUGCAUGUGCAACAGUUUCUUGCCCGCCCGCAAAAGUAGGCAAUGCUUUUUGCACUCGCAUCGAACAAAGGGAAAUUUAAUUAAA ((..(.(((((.((((.(((((((.(.....)))))))).))))))))).)..))..((((((..(....)..)))..((.((..(((....)))..)).))..)))............. ( -37.60) >DroYak_CAF1 4501 120 - 1 GGUUGGUGUUGUCAUGAGCAGCCAACAUUUUGCGGCUGCAUGUGCAACAGUUUCUUGCCCGCCCGCAAAAGUAGGCAAUGCUUUUUGCACUCGCAUCGAACAAAGGGAAAUUUAAUUAAA ((..(.(((((.((((.((((((..........)))))).))))))))).)..))..((((((..(....)..)))..((.((..(((....)))..)).))..)))............. ( -34.40) >consensus GGUUGGUGUUGUCAUGAGCAGCCGACAUUUUGCGGCUGCAUGUGCAACAGUUUCUUGCCCGCCCGCAAAAGUAGGCAAUGCUUUUUGCACUCGCAUCGAACAAAGGGAAAUUUAAUUAAA ((..(.(((((.((((.(((((((........))))))).))))))))).)..))..((((((..(....)..)))..((.((..(((....)))..)).))..)))............. (-35.93 = -36.27 + 0.33)

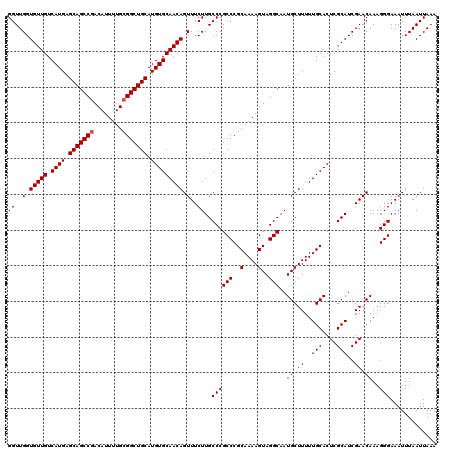
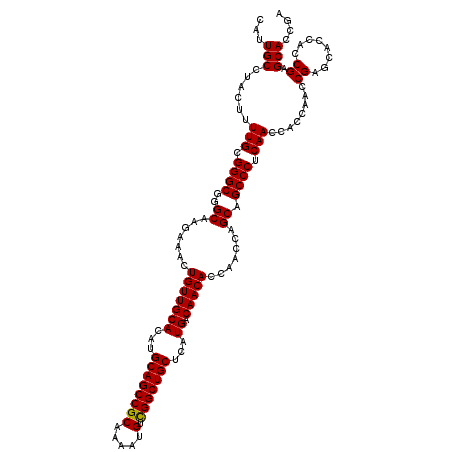
| Location | 17,468,328 – 17,468,448 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -33.31 |
| Consensus MFE | -33.53 |
| Energy contribution | -33.31 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.54 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17468328 120 + 20766785 CAUUGCCUACUUUUGCGGGCGGGCAAGAAACUGUUGCACAUGCAGCCGCAAAAUGUCGGCUGCUCAUGACAACACCAACCAGCAGCCCUCAACCACCAACCGAGCACCACCGAGCACCGA ...(((......(((.((((..((.......(((((((...((((((((.....).)))))))...)).))))).......)).)))).)))........((........)).))).... ( -33.94) >DroSim_CAF1 4273 120 + 1 CAUUGCCUACUUUUGCGGGCGGGCAAGAAACUGUUGCACAUGCAGCCGCAAAAUGUCGGCUGCUCAUGACAACACCAACCAGCAGCCCUCAACCACCAACCGAGCACCACCGAGCACCGA ...(((......(((.((((..((.......(((((((...((((((((.....).)))))))...)).))))).......)).)))).)))........((........)).))).... ( -33.94) >DroYak_CAF1 4541 118 + 1 CAUUGCCUACUUUUGCGGGCGGGCAAGAAACUGUUGCACAUGCAGCCGCAAAAUGUUGGCUGCUCAUGACAACACCAACCAGCAGCCCUCAACCACCAACCGAGCACCACCGAGCACC-- ...(((......(((.((((..((.......(((((((...((((((((.....).)))))))...)).))))).......)).)))).)))........((........)).)))..-- ( -32.04) >consensus CAUUGCCUACUUUUGCGGGCGGGCAAGAAACUGUUGCACAUGCAGCCGCAAAAUGUCGGCUGCUCAUGACAACACCAACCAGCAGCCCUCAACCACCAACCGAGCACCACCGAGCACCGA ...(((......(((.((((..((.......(((((((...((((((((.....).)))))))...)).))))).......)).)))).)))........((........)).))).... (-33.53 = -33.31 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:18 2006