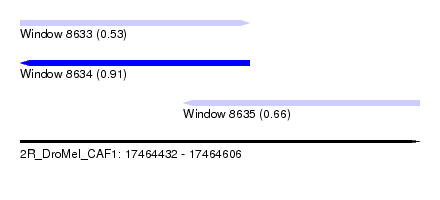
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,464,432 – 17,464,606 |
| Length | 174 |
| Max. P | 0.908698 |

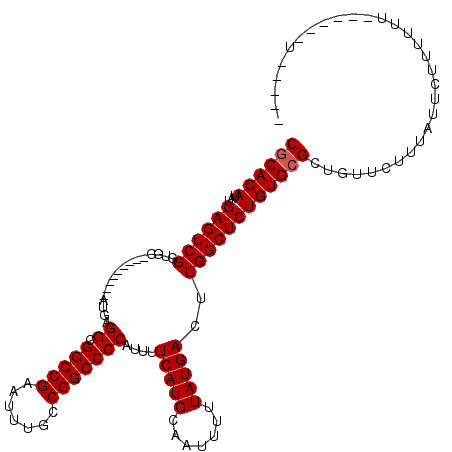
| Location | 17,464,432 – 17,464,532 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.53 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -26.13 |
| Energy contribution | -26.47 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.533793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17464432 100 + 20766785 CGCACAUAUGAGUCGGUGG---------AUGAGGCGGCCGAAUUUGCCGGCCCCAUUUUCAUGCAAUUUUUAUGACUUGGCUCUGUGCGCUGUUCUUUAUUUUUUUUU------U----- ((((((..(((((((.((.---------((((((.(((((.......)))))))....)))).)).......)))))))....))))))...................------.----- ( -29.30) >DroSim_CAF1 438 98 + 1 CGCACAUAUGAGUCGGUGG---------AAGAGGCGGCCGAAUUUGCCGGCCCCAUUUUCAUGCAAUUUUUAUGACUUGGCUCUGUGCGCUGUUCUUUAUUCUUUUU------------- ((((((...((((((((((---------(((.((.(((((.......))))))).))))))).((.......))...))))))))))))..................------------- ( -30.60) >DroYak_CAF1 456 119 + 1 C-CACAUAUGAGUCGCGGGAUGAGGCGAAUGAGGCGGCCGAAUUUGCCGGCCCCAUUUUCAUGCAAUUUUUAUGACUUGGCUCUGUGCGCUGUUCUUUAUUCUUUGUUGGGUUGUUGGCU (-((((...((((...((((...((((...(((..(((((.......)))))(((...(((((.......)))))..))))))....))))..)))).))))..)).))).......... ( -30.50) >consensus CGCACAUAUGAGUCGGUGG_________AUGAGGCGGCCGAAUUUGCCGGCCCCAUUUUCAUGCAAUUUUUAUGACUUGGCUCUGUGCGCUGUUCUUUAUUCUUUUUU______U_____ ((((((...((((((.................((.(((((.......)))))))....(((((.......)))))..))))))))))))............................... (-26.13 = -26.47 + 0.33)

| Location | 17,464,432 – 17,464,532 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.53 |
| Mean single sequence MFE | -24.63 |
| Consensus MFE | -22.63 |
| Energy contribution | -22.97 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17464432 100 - 20766785 -----A------AAAAAAAAAUAAAGAACAGCGCACAGAGCCAAGUCAUAAAAAUUGCAUGAAAAUGGGGCCGGCAAAUUCGGCCGCCUCAU---------CCACCGACUCAUAUGUGCG -----.------...................((((((......((((..........(((....))).((((((.....)))))).......---------.....))))....)))))) ( -24.90) >DroSim_CAF1 438 98 - 1 -------------AAAAAGAAUAAAGAACAGCGCACAGAGCCAAGUCAUAAAAAUUGCAUGAAAAUGGGGCCGGCAAAUUCGGCCGCCUCUU---------CCACCGACUCAUAUGUGCG -------------..................((((((......((((..........(((....))).((((((.....)))))).......---------.....))))....)))))) ( -24.90) >DroYak_CAF1 456 119 - 1 AGCCAACAACCCAACAAAGAAUAAAGAACAGCGCACAGAGCCAAGUCAUAAAAAUUGCAUGAAAAUGGGGCCGGCAAAUUCGGCCGCCUCAUUCGCCUCAUCCCGCGACUCAUAUGUG-G .................................((((((((((..((((.........))))...)))((((((.....))))))..)))..((((........))))......))))-. ( -24.10) >consensus _____A______AAAAAAGAAUAAAGAACAGCGCACAGAGCCAAGUCAUAAAAAUUGCAUGAAAAUGGGGCCGGCAAAUUCGGCCGCCUCAU_________CCACCGACUCAUAUGUGCG ...............................((((((......((((..........(((....))).((((((.....)))))).....................))))....)))))) (-22.63 = -22.97 + 0.33)

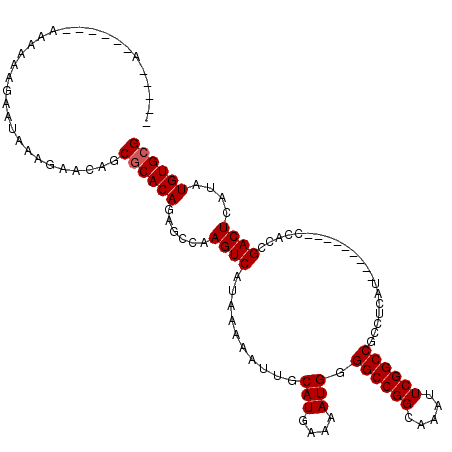
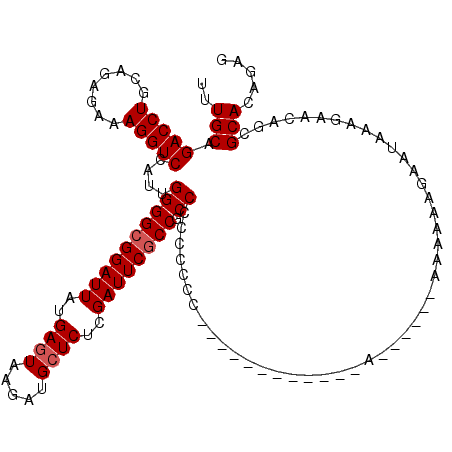
| Location | 17,464,503 – 17,464,606 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.05 |
| Mean single sequence MFE | -30.31 |
| Consensus MFE | -24.37 |
| Energy contribution | -25.03 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17464503 103 - 20766785 UUUGCAGACCUGCAGAGAAAGGUCCAUUGGGGCGGAUUAUGAGUAAGAUGCUCUCGAUUCGCCCCCCCCCCCCA-----------A------AAAAAAAAAUAAAGAACAGCGCACAGAG ..(((.(((((........)))))....((((((((((..((((.....))))..)))))))))).........-----------.------....................)))..... ( -32.30) >DroSim_CAF1 509 97 - 1 UUUGCAGACCUGCAGAGAAAGGUCCAUUGGGGCGGAUUAUGAGUAAGAUGCUCUCGAUUCGCCGCCCCCC-----------------------AAAAAGAAUAAAGAACAGCGCACAGAG ..(((.(((((........)))))....(((((((.....((((.....))))........)))))))..-----------------------...................)))..... ( -29.12) >DroYak_CAF1 535 120 - 1 UUUGCAGACCUGCUGGGAAAGGUCCAUUGGGGCGGAUUAUGAGUAAGAUGGUCCCGAUUCACCGCCGCCCCCCUUGAACAAGCCAACAACCCAACAAAGAAUAAAGAACAGCGCACAGAG ..(((.(((((........)))))((..(((((((....(((((..(......)..)))))...)))))))...))....................................)))..... ( -29.50) >consensus UUUGCAGACCUGCAGAGAAAGGUCCAUUGGGGCGGAUUAUGAGUAAGAUGCUCUCGAUUCGCCGCCCCCCCCC____________A______AAAAAAGAAUAAAGAACAGCGCACAGAG ..(((.(((((........)))))....((((((((((..((((.....))))..)))))))).))..............................................)))..... (-24.37 = -25.03 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:10 2006