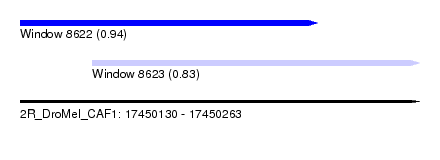
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,450,130 – 17,450,263 |
| Length | 133 |
| Max. P | 0.938057 |

| Location | 17,450,130 – 17,450,229 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 71.71 |
| Mean single sequence MFE | -24.66 |
| Consensus MFE | -11.51 |
| Energy contribution | -11.70 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17450130 99 + 20766785 UUGGGUAAUGCACUACACACACA------CACACACACUUGCCCAGCAUCCUUGCAGUCCUGCAUCCUCUCCCCCAAGUCAGCGACUUAGCG-CGUUGACUUAACU ((((((((((.............------......)).))))))))......((((....))))...........(((((((((........-))))))))).... ( -21.91) >DroVir_CAF1 186197 77 + 1 AUAGGUAAUGCACUGCCUGCAGC--------------------------UGCUGCAG--CAACUGCAAGUG-CUGCAGUCAGCGACUUUGCGACGUUGACUUAACU ..(((((......)))))(((((--------------------------.(((((((--...)))).))))-))))((((((((.(.....).))))))))..... ( -28.70) >DroSim_CAF1 169751 99 + 1 UUGGGUAAUACACUGCACACACA------CGCACACACUCGCCCAGCAUCCUUGCAGUCCUGCAUCCUUUCCCCCAAGUCAGCGACUUAGCG-CGUUGACUUAACU ((((((.......(((.......------.))).......))))))......((((....))))...........(((((((((........-))))))))).... ( -23.84) >DroYak_CAF1 150026 105 + 1 UUGGGUAAUGCACUGCACACACACACACGCAUACACACUCGCUUAGCAUCCUUGCAGUCCUGCAUCCUUUCCCCCAAGUCAGCGACUUAGCG-CGUUGACUUAACU ..(((..(((((..((............((..........))...(((....))).))..))))).....)))..(((((((((........-))))))))).... ( -24.20) >consensus UUGGGUAAUGCACUGCACACACA______CACACACACUCGCCCAGCAUCCUUGCAGUCCUGCAUCCUUUCCCCCAAGUCAGCGACUUAGCG_CGUUGACUUAACU ..(((((......))).....................................(((....))).........)).(((((((((.(.....).))))))))).... (-11.51 = -11.70 + 0.19)

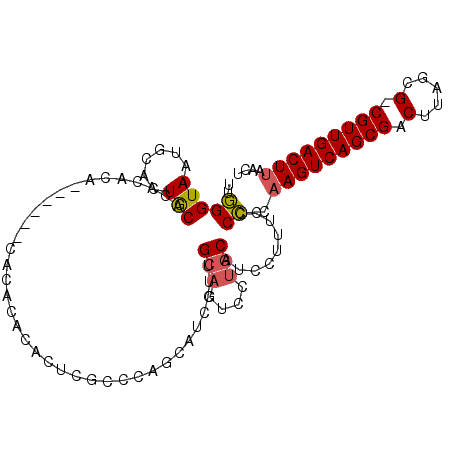
| Location | 17,450,154 – 17,450,263 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.95 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -16.45 |
| Energy contribution | -17.03 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17450154 109 + 20766785 ACACACACUUGCCCAGCAUCCUUGC----AGUCCUGCAUCCUCUCCCCCAAGUCAGCGACUUAGCG-CGUUGACUUAACUUGGCAGGGAUUUACGACAACGCUCCACGGAGCUC------ .......((((((.((......(((----(....))))...........(((((((((........-)))))))))..)).)))))).............((((....))))..------ ( -32.20) >DroVir_CAF1 186220 94 + 1 -------------------UGCUGC----AG--CAACUGCAAGUG-CUGCAGUCAGCGACUUUGCGACGUUGACUUAACUUGGCAGGGAUUUACGGCAACGCUUCACGGAGCUGCGACAC -------------------((.(((----((--(..((((((((.-....((((((((.(.....).))))))))..)))).)))).((....((....))..)).....)))))).)). ( -31.70) >DroGri_CAF1 181823 86 + 1 -------------------UGUUGC----AA--GUGCGGCUGCUG-CUGCAGUCAGCGACUUUGCGACGUUGACUUAACUUGGCAGGGAUUUACAGCCACGCUUCGCGGAGC-------- -------------------..((((----((--(((.((((((((-(((.((((((((.(.....).)))))))).....)))))).......))))).))))).))))...-------- ( -31.01) >DroEre_CAF1 163691 109 + 1 ACACACACUGGCCCUGCAUCCUUGC----AGUCCUGCAUCCUUUCCCCCAAGUCAGCGACUUAGCG-CGUUGACUUAACUUGGCAGGGAUUUACGACAACGCUCCACGGAGCUC------ ...........((((((.....(((----(....))))...........(((((((((........-)))))))))......))))))............((((....))))..------ ( -34.20) >DroYak_CAF1 150056 109 + 1 AUACACACUCGCUUAGCAUCCUUGC----AGUCCUGCAUCCUUUCCCCCAAGUCAGCGACUUAGCG-CGUUGACUUAACUUGGCAGGGAUUUACGACAACGCUCCACGGAGCUC------ ...............(((....)))----.(((.((.((((((.((...(((((((((........-))))))))).....)).)))))).)).)))...((((....))))..------ ( -32.90) >DroMoj_CAF1 229830 91 + 1 -C-----------------UGCUGCCUGCUG-CCUGCUGCCUG----UGCAGUCAGCGACUUUGCGACGUUGACUUAACUUGGCAGGGAUUUACGGCAACGCUUCACGGAGCUU------ -.-----------------.(.(((((((((-.((((......----.)))).))))).((((((.(.((((...)))).).))))))......)))).)((((....))))..------ ( -31.50) >consensus _C_________________CCUUGC____AG_CCUGCAGCCUCUC_CCCAAGUCAGCGACUUAGCG_CGUUGACUUAACUUGGCAGGGAUUUACGACAACGCUCCACGGAGCUC______ ................................(((((.............((((((((.(.....).)))))))).......))))).............((((....))))........ (-16.45 = -17.03 + 0.58)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:59 2006