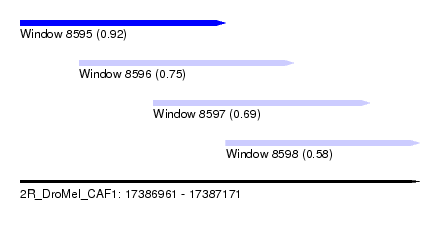
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,386,961 – 17,387,171 |
| Length | 210 |
| Max. P | 0.918060 |

| Location | 17,386,961 – 17,387,069 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 81.71 |
| Mean single sequence MFE | -36.28 |
| Consensus MFE | -26.12 |
| Energy contribution | -28.23 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918060 |
| Prediction | RNA |
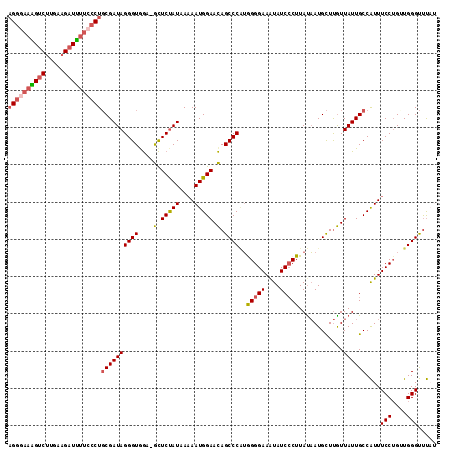
Download alignment: ClustalW | MAF
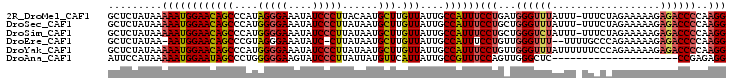
>2R_DroMel_CAF1 17386961 108 + 20766785 UUC----UUCGUUUCUCGGGUUUUCGCCUGCACCUGUUGUCGCACCAGUAAACUGGAAAAGUUGCUGAAAACUAAGGGAAAGUCUUGAAGAUUUUACCUGCGAUAGGGUAGA .((----((((((((((.(((((((((.(((..(....)..)))((((....)))).......)).)))))))..))))))....)))))).(((((((......))))))) ( -30.60) >DroSec_CAF1 102830 108 + 1 UUC----UUCCAUUCUCGGGUUUUCGCCUGCACCUGUUGUCGCACCAGUAAACUGGAAAAGUUGCUGAAAAGUAAGGGAAGGUCUUGAAGAUUUUCCCUGCGAUAGGGUGGA ...----.(((((((.(((((....))))).......(((((((((((....)))).....(((((....)))))(((((((((.....))))))))))))))))))))))) ( -42.30) >DroSim_CAF1 106273 108 + 1 UUC----UUCCUUUCUCGGGUUUUCGCCUGCACCUGUUGUCGCACCAGUAAACUGGAAAAGUUGCUGAAAAGUAUGGGAAGGUCUUGAAGAUUUUCCCUGCGAUAGGGUGGA ...----.........(((((....)))))(((((..(((((((((((....))))......((((....)))).(((((((((.....))))))))))))))))))))).. ( -40.20) >DroEre_CAF1 101663 107 + 1 UUC----UUGCUUU-CCGGGUUUUCGCCUGCACCUGUUGUCGCACCAGUAAACUGGAAAAGUUGCUGAAAAGUAAGGGAAAGUCUUGAAGAUUUUCCCUGCGAUAGGGUGGA ...----.......-.(((((....)))))(((((..(((((((((((....)))).....(((((....)))))(((((((((.....))))))))))))))))))))).. ( -42.10) >DroYak_CAF1 85078 112 + 1 UUUUGUAUUCCUUUCCCGGAUUUGCGCCUGCACCUGUUGUCGCACCAGUAAACUGGAAAAGUUGCCGAAAAGUGAGGGAAAGUCUUGAAGAUUUUCCCUGCGAUAGGGUGGA ....(((.(((......)))..))).....(((((..((((((.((((....))))..................((((((((((.....))))))))))))))))))))).. ( -38.70) >DroPer_CAF1 109839 97 + 1 ---------CAAAACACACAUCUCCACCUGCACCUGUUGUCGCACCAG-AAACUGGAAAACUGGCCGAAAAUUCGGCACA-----AGAAAAUGCCCCAUUCGAUGAGAGGUA ---------......................((((.(..(((..((((-...))))....((.(((((....)))))...-----)).............)))..).)))). ( -23.80) >consensus UUC____UUCCUUUCUCGGGUUUUCGCCUGCACCUGUUGUCGCACCAGUAAACUGGAAAAGUUGCUGAAAAGUAAGGGAAAGUCUUGAAGAUUUUCCCUGCGAUAGGGUGGA ................(((((....)))))(((((..(((((((((((....))))...................(((((((((.....))))))))))))))))))))).. (-26.12 = -28.23 + 2.11)

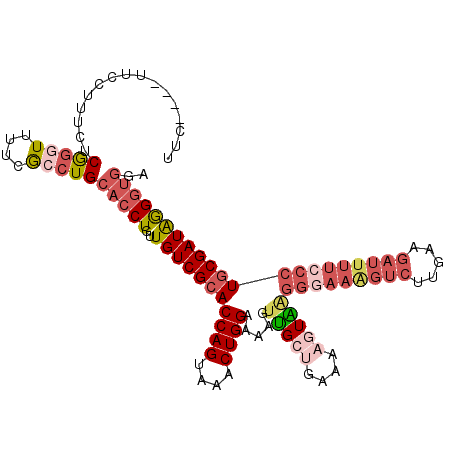
| Location | 17,386,992 – 17,387,105 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 85.19 |
| Mean single sequence MFE | -34.63 |
| Consensus MFE | -24.87 |
| Energy contribution | -26.65 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17386992 113 + 20766785 GUUGUCGCACCAGUAAACUGGAAAAGUUGCUGAAAACUAAGGGAAAGUCUUGAAGAUUUUACCUGCGAUAGGGUAGAGCUCUAUAAAA----AUGGAACAGCCCAUAGGGAAAUAUC ..((((((.((((....))))...((((......)))).(((.((((((.....)))))).)))))))))((((.(...(((((....----))))).).))))............. ( -30.00) >DroSec_CAF1 102861 113 + 1 GUUGUCGCACCAGUAAACUGGAAAAGUUGCUGAAAAGUAAGGGAAGGUCUUGAAGAUUUUCCCUGCGAUAGGGUGGAGCUCUAUAAAA----AUGGAACAGCCCAUGGGGAAAUAUC ..(((((((((((....)))).....(((((....)))))(((((((((.....))))))))))))))))((((...(.(((((....----))))).).))))............. ( -39.40) >DroSim_CAF1 106304 113 + 1 GUUGUCGCACCAGUAAACUGGAAAAGUUGCUGAAAAGUAUGGGAAGGUCUUGAAGAUUUUCCCUGCGAUAGGGUGGAGCUCUAUAAAA----AUGGAACAGCCCAUGGGGAAAUAUC ..(((((((((((....))))......((((....)))).(((((((((.....))))))))))))))))((((...(.(((((....----))))).).))))............. ( -38.10) >DroEre_CAF1 101693 112 + 1 GUUGUCGCACCAGUAAACUGGAAAAGUUGCUGAAAAGUAAGGGAAAGUCUUGAAGAUUUUCCCUGCGAUAGGGUGGAGCUCUAUAA-A----AUGGAACAGCCCGUAGGGAAAUAUC ..(((((((((((....)))).....(((((....)))))(((((((((.....))))))))))))))))((((...(.(((((..-.----))))).).))))............. ( -38.70) >DroYak_CAF1 85113 113 + 1 GUUGUCGCACCAGUAAACUGGAAAAGUUGCCGAAAAGUGAGGGAAAGUCUUGAAGAUUUUCCCUGCGAUAGGGUGGAGCUCUAUAAAA----AUGGAACAGCCCAUGGGGAAAUAUC ..((((((.((((....))))..................((((((((((.....))))))))))))))))((((...(.(((((....----))))).).))))............. ( -36.70) >DroPer_CAF1 109865 107 + 1 GUUGUCGCACCAG-AAACUGGAAAACUGGCCGAAAAUUCGGCACA-----AGAAAAUGCCCCAUUCGAUGAGAGGUAGCCCUAUAAAAUUCCAUGGAAUUGCCCUAAGGGAGA---- ......(((((((-...))))....((.(((((....)))))...-----))....)))(((..((.....))(((((..((((........))))..)))))....)))...---- ( -24.90) >consensus GUUGUCGCACCAGUAAACUGGAAAAGUUGCUGAAAAGUAAGGGAAAGUCUUGAAGAUUUUCCCUGCGAUAGGGUGGAGCUCUAUAAAA____AUGGAACAGCCCAUAGGGAAAUAUC .((((((((((((....))))...................(((((((((.....)))))))))))))))))((((...((((..........((((......))))))))...)))) (-24.87 = -26.65 + 1.78)

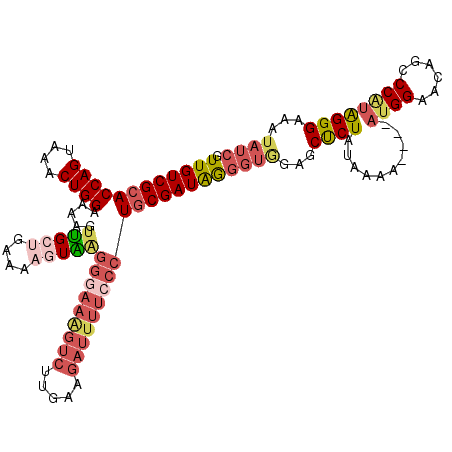

| Location | 17,387,031 – 17,387,145 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 86.40 |
| Mean single sequence MFE | -34.77 |
| Consensus MFE | -27.43 |
| Energy contribution | -27.85 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17387031 114 + 20766785 AGGGAAAGUCUUGAAGAUUUUACCUGCGAUAGGGUAGA-GCUCUAUAAAAAUGGAACAGCCCAUAGGGAAAUAUCCCUUACAAUGCUUGUUAUUGCCAUUUCCUGAUGGGUUUAU .(((((((((.....)))))).)))...((((((....-.))))))...........(((((((.(((((((.......(((.....))).......))))))).)))))))... ( -29.34) >DroSec_CAF1 102900 114 + 1 AGGGAAGGUCUUGAAGAUUUUCCCUGCGAUAGGGUGGA-GCUCUAUAAAAAUGGAACAGCCCAUGGGGAAAUAUCCCUUAUAAUGCUUGUUAUUGCCAUUUCCUGCUGGGUUUAU ((((((((((.....))))))))))((((((((((...-(.(((((....))))).).))))..(((((....)))))............))))))....(((....)))..... ( -34.60) >DroSim_CAF1 106343 114 + 1 UGGGAAGGUCUUGAAGAUUUUCCCUGCGAUAGGGUGGA-GCUCUAUAAAAAUGGAACAGCCCAUGGGGAAAUAUCCCUUAUAAUGCUUGUUAUUGCCAUUUCCUGCUGGGUCUAU .(((((((((.....))))))))).((((((((((...-(.(((((....))))).).))))..(((((....)))))............))))))....(((....)))..... ( -33.50) >DroEre_CAF1 101732 110 + 1 AGGGAAAGUCUUGAAGAUUUUCCCUGCGAUAGGGUGGA-GCUCUAUAA-AAUGGAACAGCCCGUAGGGAAAUAUC-CUUAUAAUGCUUGUUAUUGCCAUUUCCUGUUGGGUUU-- ((((((((((.....)))))))))).(((((((((((.-...)))).(-(((((((((((..(((((((....))-)))))...)).))))....))))))))))))).....-- ( -37.80) >DroYak_CAF1 85152 114 + 1 AGGGAAAGUCUUGAAGAUUUUCCCUGCGAUAGGGUGGA-GCUCUAUAAAAAUGGAACAGCCCAUGGGGAAAUAUCCCUUAUAAUGCUUGUUAUUGCCAUUUCCUGUUGGGUUUAU ((((((((((.....)))))))))).(((((((((((.-...))))..((((((((((((....(((((....)))))......)).))))....)))))))))))))....... ( -37.30) >DroAna_CAF1 93772 110 + 1 AGC--CUGCCCGAGAGAUGUGCCCU-CGAUAGGGUAAAAAUUCCAUAAAAAUGGAAUAGCCCUGGGGGAAGUAUCCCUUAUUAUGUUCAUUAUUGCCGUUUCCAGUUGGGCUC-- ...--..((((((((((((.((...-...((((((....(((((((....))))))).))))))(((((....)))))................))))))))...))))))..-- ( -36.10) >consensus AGGGAAAGUCUUGAAGAUUUUCCCUGCGAUAGGGUGGA_GCUCUAUAAAAAUGGAACAGCCCAUGGGGAAAUAUCCCUUAUAAUGCUUGUUAUUGCCAUUUCCUGUUGGGUUUAU ((((((((((.....))))))))))((((((((((....(.(((((....))))).).))))..(((((....)))))............))))))....(((....)))..... (-27.43 = -27.85 + 0.42)

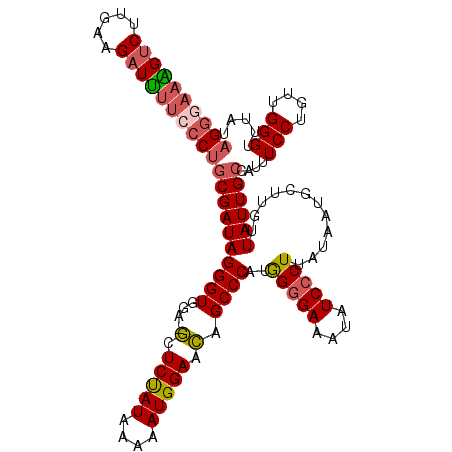
| Location | 17,387,069 – 17,387,171 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 82.89 |
| Mean single sequence MFE | -25.63 |
| Consensus MFE | -17.04 |
| Energy contribution | -17.32 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17387069 102 + 20766785 GCUCUAUAAAAAUGGAACAGCCCAUAGGGAAAUAUCCCUUACAAUGCUUGUUAUUGCCAUUUCCUGAUGGGUUUAUUU-UUUCUAGAAAAAGAGACCCCAAGG .........((((((((((((....(((((....)))))......)).))))....))))))(((...((((((.(((-(((....))))))))))))..))) ( -24.90) >DroSec_CAF1 102938 102 + 1 GCUCUAUAAAAAUGGAACAGCCCAUGGGGAAAUAUCCCUUAUAAUGCUUGUUAUUGCCAUUUCCUGCUGGGUUUAUUU-UUUCUAGAAAAAGAGACCCCAAGG .........((((((((((((....(((((....)))))......)).))))....))))))(((...((((((.(((-(((....))))))))))))..))) ( -24.90) >DroSim_CAF1 106381 102 + 1 GCUCUAUAAAAAUGGAACAGCCCAUGGGGAAAUAUCCCUUAUAAUGCUUGUUAUUGCCAUUUCCUGCUGGGUCUAUUU-UUUCUAGAAAAAGAGACCCCAAGG .........((((((((((((....(((((....)))))......)).))))....))))))(((...((((((.(((-(((....))))))))))))..))) ( -27.50) >DroEre_CAF1 101770 99 + 1 GCUCUAUAA-AAUGGAACAGCCCGUAGGGAAAUAUC-CUUAUAAUGCUUGUUAUUGCCAUUUCCUGUUGGGUUU--UUUUGCCCAGAAAAAGAGACCCCAAGG ........(-(((((((((((..(((((((....))-)))))...)).))))....))))))(((...((((((--((((........))))))))))..))) ( -27.60) >DroYak_CAF1 85190 103 + 1 GCUCUAUAAAAAUGGAACAGCCCAUGGGGAAAUAUCCCUUAUAAUGCUUGUUAUUGCCAUUUCCUGUUGGGUUUAUUUUUUCCCAGAAAAAGAGACCCCAAGG .........((((((((((((....(((((....)))))......)).))))....))))))(((...((((((.(((((((...)))))))))))))..))) ( -26.00) >DroAna_CAF1 93808 82 + 1 AUUCCAUAAAAAUGGAAUAGCCCUGGGGGAAGUAUCCCUUAUUAUGUUCAUUAUUGCCGUUUCCAGUUGGGCUC---------------------CCGAGAGG (((((((....)))))))((((((((((((....)))))))...........((((.......)))).))))).---------------------((....)) ( -22.90) >consensus GCUCUAUAAAAAUGGAACAGCCCAUGGGGAAAUAUCCCUUAUAAUGCUUGUUAUUGCCAUUUCCUGUUGGGUUUAUUU_UUUCUAGAAAAAGAGACCCCAAGG .........((((((((((((....(((((....)))))......))).)))....))))))(((...((((((..................))))))..))) (-17.04 = -17.32 + 0.28)
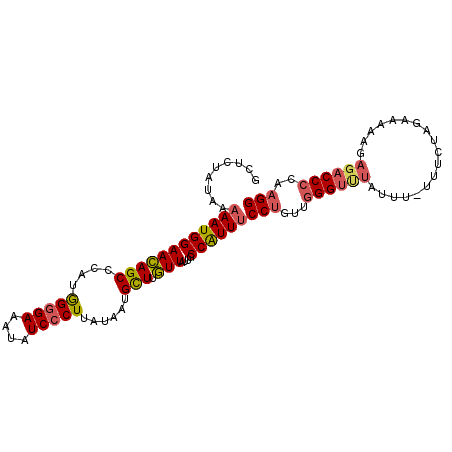
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:35 2006