
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,386,739 – 17,386,830 |
| Length | 91 |
| Max. P | 0.984166 |

| Location | 17,386,739 – 17,386,830 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 79.67 |
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -15.92 |
| Energy contribution | -18.08 |
| Covariance contribution | 2.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
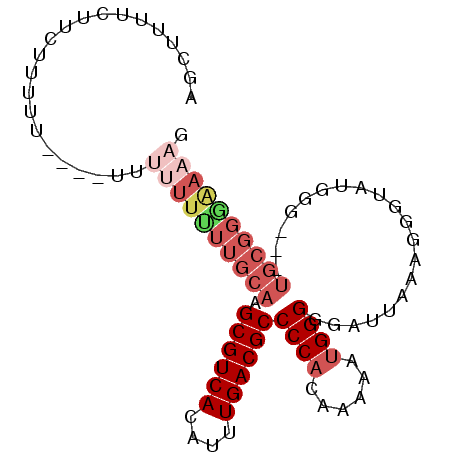
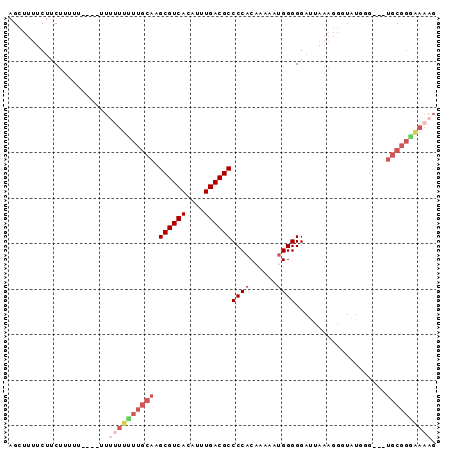
>2R_DroMel_CAF1 17386739 91 + 20766785 AGCUUUUCUUCUUUUUCUUUUUUUUUUUUGCAAGCGUCACAUUUGACGCCCCACAAAAAUGGGGGAUUAAAGGGUAUGGG---UGCGGAAAAAG ......................((((((((((..(((.((.(((((..(((((......)))))..)))))..)))))..---)))))))))). ( -22.90) >DroSec_CAF1 102616 83 + 1 AGCUUUUCUUCUUU------UU--UUUUUGCAAGCGUCACAUUUGACGCCCCACAAAAAUGGGGGAUUAAAGGGUAUGGG---UGCGGGAAAAG ..............------((--((((((((..(((.((.(((((..(((((......)))))..)))))..)))))..---)))))))))). ( -22.50) >DroSim_CAF1 106057 85 + 1 AGCUUUUCUUCUUU------UUUUUUUUUGCAAGCGUCACAUUUGACGCCCCACAAAAAUGGGGGAUUAAAGGGUAUGGG---UGCGGGAAAAG ..............------..((((((((((..(((.((.(((((..(((((......)))))..)))))..)))))..---)))))))))). ( -22.50) >DroEre_CAF1 101464 69 + 1 AGCUUUUCUCCUUUUUUUUUUUUUUUGUUGCAAGCGUCACAUUUGACGCCCCACAAAAAGGGGGGUUUU------------------------- ......(((((((((((.......(((...)))((((((....)))))).....)))))))))))....------------------------- ( -16.20) >DroYak_CAF1 84845 91 + 1 AGCUUUUCUCCUUUUUU---UUUUUUGUUGCAAGCGUCACAUUUGACGCCCCACCAAAAUGGGGGAUUAAAGGGUGUGGGGGUUGCGGCGAAAG .................---..(((((((((((.(.((((((((((..(((((......)))))..))))...)))))).).))))))))))). ( -31.30) >consensus AGCUUUUCUUCUUUUU____UUUUUUUUUGCAAGCGUCACAUUUGACGCCCCACAAAAAUGGGGGAUUAAAGGGUAUGGG___UGCGGGAAAAG ......................((((((((((.((((((....))))))((((......))))....................)))))))))). (-15.92 = -18.08 + 2.16)

| Location | 17,386,739 – 17,386,830 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 79.67 |
| Mean single sequence MFE | -19.06 |
| Consensus MFE | -14.88 |
| Energy contribution | -15.68 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
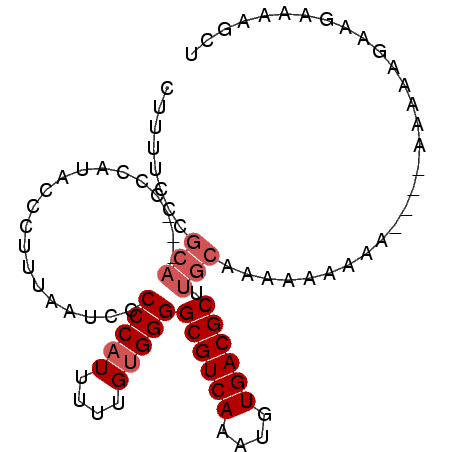
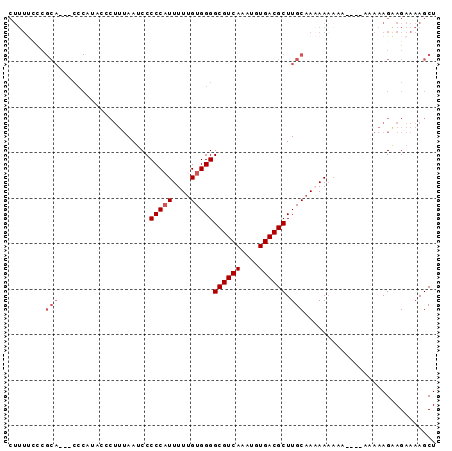
>2R_DroMel_CAF1 17386739 91 - 20766785 CUUUUUCCGCA---CCCAUACCCUUUAAUCCCCCAUUUUUGUGGGGCGUCAAAUGUGACGCUUGCAAAAAAAAAAAAGAAAAAGAAGAAAAGCU (((((((....---.................(((((....)))))((((((....))))))................))))))).......... ( -20.90) >DroSec_CAF1 102616 83 - 1 CUUUUCCCGCA---CCCAUACCCUUUAAUCCCCCAUUUUUGUGGGGCGUCAAAUGUGACGCUUGCAAAAA--AA------AAAGAAGAAAAGCU ((((((.....---.................(((((....)))))((((((....)))))).........--..------......)))))).. ( -19.30) >DroSim_CAF1 106057 85 - 1 CUUUUCCCGCA---CCCAUACCCUUUAAUCCCCCAUUUUUGUGGGGCGUCAAAUGUGACGCUUGCAAAAAAAAA------AAAGAAGAAAAGCU ((((((.....---.................(((((....)))))((((((....)))))).............------......)))))).. ( -19.30) >DroEre_CAF1 101464 69 - 1 -------------------------AAAACCCCCCUUUUUGUGGGGCGUCAAAUGUGACGCUUGCAACAAAAAAAAAAAAAAAGGAGAAAAGCU -------------------------..........(((((((.((((((((....))))))))...)))))))..................... ( -15.10) >DroYak_CAF1 84845 91 - 1 CUUUCGCCGCAACCCCCACACCCUUUAAUCCCCCAUUUUGGUGGGGCGUCAAAUGUGACGCUUGCAACAAAAAA---AAAAAAGGAGAAAAGCU .((((.((.....................(((((.....)).)))((((((....)))))).............---......)).)))).... ( -20.70) >consensus CUUUUCCCGCA___CCCAUACCCUUUAAUCCCCCAUUUUUGUGGGGCGUCAAAUGUGACGCUUGCAAAAAAAAA____AAAAAGAAGAAAAGCU ........(((....................(((((....)))))((((((....)))))).)))............................. (-14.88 = -15.68 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:32 2006