| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,360,366 – 17,360,460 |
| Length | 94 |
| Max. P | 0.957033 |

| Location | 17,360,366 – 17,360,460 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.76 |
| Mean single sequence MFE | -26.52 |
| Consensus MFE | -15.61 |
| Energy contribution | -16.00 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.957033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
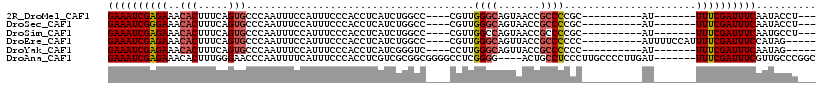
>2R_DroMel_CAF1 17360366 94 + 20766785 GAAAUCGAGAAACACUUUCAGUGCCCAAUUUCCAUUUCCCACCUCAUCUGGCC----CGUUGGGCAGUAACCGCCCCGC----------AU-------UUUCGAUUUCAAUACCU--- (((((((((((........(.((((((((..(((..............)))..----.)))))))).)....((...))----------.)-------)))))))))).......--- ( -25.94) >DroSec_CAF1 76206 94 + 1 GAAAUCGGGAAACACUUUCAGUGCCCAAUUUCCAUUUCCCACCUCAUCUGGCC----CGUUGGGCAGUAACCGCCCCGC----------AU-------UUUCGAUUUCAAUACCU--- (((((((..((........(.((((((((..(((..............)))..----.)))))))).)....((...))----------.)-------)..))))))).......--- ( -25.14) >DroSim_CAF1 78750 94 + 1 GAAAUCGAGAAACACUUUCAGUGCCCAAUUUCCAUUUCCCACCUCAUCUGGCC----CGUUGGCCAGUAACCGCCCCGC----------AU-------UUUCGAUUUCAAUGCCU--- (((((((((((.(((.....)))........................((((((----....))))))............----------.)-------)))))))))).......--- ( -24.40) >DroEre_CAF1 74829 99 + 1 GAAAUCGAGAAACACUUUCAGUGCCCAAUUUCCAUUUCCCACCUCAUCUGGCC----CGUUGGGCAGUUACCGCCCCCC----------AUUUUCCAUUUUCGAUUUCCAUAG----- (((((((((((..........((((((((..(((..............)))..----.))))))))((....)).....----------........))))))))))).....----- ( -22.94) >DroYak_CAF1 56601 92 + 1 GAAAUCGAGAAACACUUUCAGUGCCCAAUUUCCAUUUCCCACCUCAUCGGGUC----CCUUGGGCAGUUACCGCCCCCC----------AU-------UUUCGAUUUCAAUAG----- (((((((((((.(((.....))).........................(((..----....((((.......)))))))----------.)-------)))))))))).....----- ( -24.10) >DroAna_CAF1 69585 107 + 1 GAAAUCGAGAAACACUUUGGGAACCCAAUUUUCAUUUCCCACCUCGUCGCGGCGGGGCCUCGGGG----ACUGCCUCCCUUGCCCCUUGAU-------UUUCGAUUUCGUUGCCCGGC (((((((((((.((...((((((............))))))....((....))(((((...((((----(.....))))).))))).)).)-------)))))))))).......... ( -36.60) >consensus GAAAUCGAGAAACACUUUCAGUGCCCAAUUUCCAUUUCCCACCUCAUCUGGCC____CGUUGGGCAGUAACCGCCCCCC__________AU_______UUUCGAUUUCAAUACCU___ ((((((((((..(((.....)))......................................((((.......))))......................)))))))))).......... (-15.61 = -16.00 + 0.39)
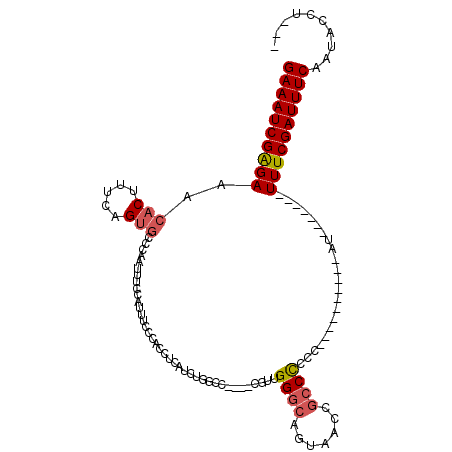
| Location | 17,360,366 – 17,360,460 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.76 |
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -17.19 |
| Energy contribution | -18.08 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17360366 94 - 20766785 ---AGGUAUUGAAAUCGAAA-------AU----------GCGGGGCGGUUACUGCCCAACG----GGCCAGAUGAGGUGGGAAAUGGAAAUUGGGCACUGAAAGUGUUUCUCGAUUUC ---.......((((((((((-------((----------((.((((((...))))))..((----((((.(((................))).))).)))...)))))).)))))))) ( -28.79) >DroSec_CAF1 76206 94 - 1 ---AGGUAUUGAAAUCGAAA-------AU----------GCGGGGCGGUUACUGCCCAACG----GGCCAGAUGAGGUGGGAAAUGGAAAUUGGGCACUGAAAGUGUUUCCCGAUUUC ---.......((((((....-------.(----------(..((((((...))))))..))----.(((......)))(((((((....))).(((((.....))))))))))))))) ( -27.50) >DroSim_CAF1 78750 94 - 1 ---AGGCAUUGAAAUCGAAA-------AU----------GCGGGGCGGUUACUGGCCAACG----GGCCAGAUGAGGUGGGAAAUGGAAAUUGGGCACUGAAAGUGUUUCUCGAUUUC ---.......((((((((((-------((----------((..........((((((....----))))))....((((...(((....)))...))))....)))))).)))))))) ( -28.50) >DroEre_CAF1 74829 99 - 1 -----CUAUGGAAAUCGAAAAUGGAAAAU----------GGGGGGCGGUAACUGCCCAACG----GGCCAGAUGAGGUGGGAAAUGGAAAUUGGGCACUGAAAGUGUUUCUCGAUUUC -----.....((((((((...(((....(----------(..((((((...))))))..))----..))).....((((...(((....)))...))))...........)))))))) ( -28.20) >DroYak_CAF1 56601 92 - 1 -----CUAUUGAAAUCGAAA-------AU----------GGGGGGCGGUAACUGCCCAAGG----GACCCGAUGAGGUGGGAAAUGGAAAUUGGGCACUGAAAGUGUUUCUCGAUUUC -----.....((((((((..-------..----------...((((((...))))))..(.----.((.(.....((((...(((....)))...))))....).))..))))))))) ( -27.10) >DroAna_CAF1 69585 107 - 1 GCCGGGCAACGAAAUCGAAA-------AUCAAGGGGCAAGGGAGGCAGU----CCCCGAGGCCCCGCCGCGACGAGGUGGGAAAUGAAAAUUGGGUUCCCAAAGUGUUUCUCGAUUUC ....(....)((((((((..-------.....(((((..(((.......----.)))...))))).((((......))))((((((....((((....))))..)))))))))))))) ( -38.50) >consensus ___AGGUAUUGAAAUCGAAA_______AU__________GCGGGGCGGUUACUGCCCAACG____GGCCAGAUGAGGUGGGAAAUGGAAAUUGGGCACUGAAAGUGUUUCUCGAUUUC ..........((((((((........................((((((...))))))..................((((...(((....)))...))))...........)))))))) (-17.19 = -18.08 + 0.89)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:26 2006