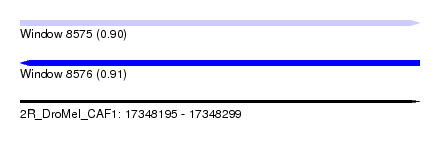
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,348,195 – 17,348,299 |
| Length | 104 |
| Max. P | 0.906752 |

| Location | 17,348,195 – 17,348,299 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.34 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -15.58 |
| Energy contribution | -15.92 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
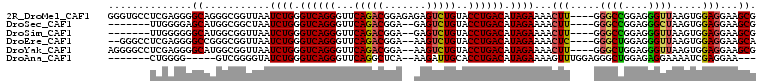
>2R_DroMel_CAF1 17348195 104 + 20766785 CGCUUCCUCCACUUAACCCUCCGGCCC----AAGUUUUCUAUGUCAGGUACAGACUCUCUCCGUCUGAACCCUGACCCAGAUUAACCGCCCUGCCCCUCGAGGCACCC ......................(((..----..(((.(((..((((((..(((((.......)))))...))))))..)))..))).))).((((......))))... ( -25.90) >DroSec_CAF1 64086 95 + 1 CGCUUCCUCCACUUAGCCCUCCGGCCC----AAGUUUUCUAUGUCAGGUACAGACUC--UCCGUCUGAACCCUGACCCAGAUUAGCCGCCAUGCUCCCCAA------- ..............(((....((((..----......(((..((((((..(((((..--...)))))...))))))..)))...))))....)))......------- ( -23.80) >DroSim_CAF1 66526 95 + 1 CGCUUCCUCCACUUAACCCUCCGGCCC----AAGUUUUCUAUGUCAGGUACAGACUC--UCCGUCUGAACCCUGACCCAGAUUAACCGCCAUGCCCCCCAA------- ......................(((..----..(((.(((..((((((..(((((..--...)))))...))))))..)))..))).)))...........------- ( -22.60) >DroEre_CAF1 62669 100 + 1 UGCUUCCUCCACUUAACCCUCCAGCCC----GAGUUUUCUAUGUCAGGUACAGACUU--UCCGUCUGAACCCUGACCCAGAUUAACCGCCCGGCCCCUCGAGGCCC-- .......................((((----(((...(((..((((((..(((((..--...)))))...))))))..)))....((....))...)))).)))..-- ( -28.10) >DroYak_CAF1 43906 102 + 1 CGCUUCCUCCACUUAACCCUCCAGCCC----AAGUUUUCUAUGUCAGGUACAGACUU--UCCGUCUGAACCCUGACCCAGAUUAACCGCCAUGCCCCUCGAGGCCCCU .(((((.....................----..(((.(((..((((((..(((((..--...)))))...))))))..)))..))).((...)).....))))).... ( -23.10) >DroAna_CAF1 57126 91 + 1 ---UUCCUCGAUUUUCCUCUCCAGCCCUCCAAACUUUUCUAUGUCAGGUGCAAUCUU--UGAGCCUGAACCCUGACCCAGAUACCCCGAC-----CCCCAG------- ---....(((.................................((((((.((.....--)).))))))...(((...)))......))).-----......------- ( -10.20) >consensus CGCUUCCUCCACUUAACCCUCCAGCCC____AAGUUUUCUAUGUCAGGUACAGACUC__UCCGUCUGAACCCUGACCCAGAUUAACCGCCAUGCCCCCCAA_______ .....................................(((..((((((..(((((.......)))))...))))))..)))........................... (-15.58 = -15.92 + 0.33)


| Location | 17,348,195 – 17,348,299 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 81.34 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -24.71 |
| Energy contribution | -24.68 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
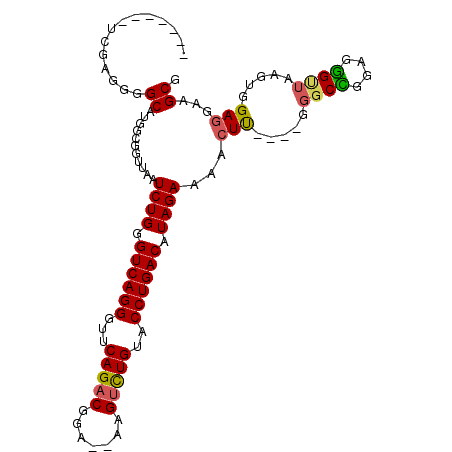
>2R_DroMel_CAF1 17348195 104 - 20766785 GGGUGCCUCGAGGGGCAGGGCGGUUAAUCUGGGUCAGGGUUCAGACGGAGAGAGUCUGUACCUGACAUAGAAAACUU----GGGCCGGAGGGUUAAGUGGAGGAAGCG ...(((((....)))))..((......((((.((((((...(((((.......)))))..)))))).))))..((((----((.((...)).)))))).......)). ( -37.40) >DroSec_CAF1 64086 95 - 1 -------UUGGGGAGCAUGGCGGCUAAUCUGGGUCAGGGUUCAGACGGA--GAGUCUGUACCUGACAUAGAAAACUU----GGGCCGGAGGGCUAAGUGGAGGAAGCG -------............((..((..((((.((((((...(((((...--..)))))..)))))).))))..((((----((.((...)).))))))...))..)). ( -34.10) >DroSim_CAF1 66526 95 - 1 -------UUGGGGGGCAUGGCGGUUAAUCUGGGUCAGGGUUCAGACGGA--GAGUCUGUACCUGACAUAGAAAACUU----GGGCCGGAGGGUUAAGUGGAGGAAGCG -------............((......((((.((((((...(((((...--..)))))..)))))).))))..((((----((.((...)).)))))).......)). ( -30.20) >DroEre_CAF1 62669 100 - 1 --GGGCCUCGAGGGGCCGGGCGGUUAAUCUGGGUCAGGGUUCAGACGGA--AAGUCUGUACCUGACAUAGAAAACUC----GGGCUGGAGGGUUAAGUGGAGGAAGCA --.((((.((((.((((....))))..((((.((((((...(((((...--..)))))..)))))).))))...)))----)))))...................... ( -38.30) >DroYak_CAF1 43906 102 - 1 AGGGGCCUCGAGGGGCAUGGCGGUUAAUCUGGGUCAGGGUUCAGACGGA--AAGUCUGUACCUGACAUAGAAAACUU----GGGCUGGAGGGUUAAGUGGAGGAAGCG ....((((....))))...((......((((.((((((...(((((...--..)))))..)))))).))))..((((----((.(.....).)))))).......)). ( -35.40) >DroAna_CAF1 57126 91 - 1 -------CUGGGG-----GUCGGGGUAUCUGGGUCAGGGUUCAGGCUCA--AAGAUUGCACCUGACAUAGAAAAGUUUGGAGGGCUGGAGAGGAAAAUCGAGGAA--- -------(((...-----(((((((((((((((((........))))).--.))).))).)))))).)))..............((.((........)).))...--- ( -21.90) >consensus _______UCGAGGGGCAUGGCGGUUAAUCUGGGUCAGGGUUCAGACGGA__AAGUCUGUACCUGACAUAGAAAACUU____GGGCCGGAGGGUUAAGUGGAGGAAGCG ..............((...........((((.((((((...(((((.......)))))..)))))).))))...(((.....((((....)))).....)))...)). (-24.71 = -24.68 + -0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:15 2006