
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,297,722 – 17,297,906 |
| Length | 184 |
| Max. P | 0.983561 |

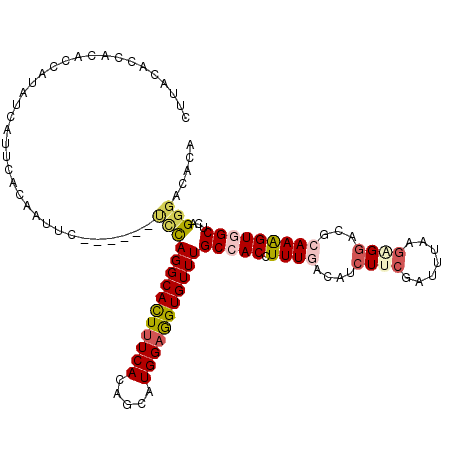
| Location | 17,297,722 – 17,297,833 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.29 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -17.11 |
| Energy contribution | -17.75 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17297722 111 - 20766785 UUUACACUACACCAUAUCAUUCACAAUUC------UCUAGGCACUUUCACAGCAUGGAGGUGUUUGCCACCUUUGAUAUCUUCAAUUUAAGAGGAAUCAAAGUGGCUCAGGGACACA ...........................((------((((((((((((((.....)))))))))))(((((.((((((.(((((.......))))))))))))))))..))))).... ( -32.70) >DroGri_CAF1 302 109 - 1 CUUAAGAUAUAAUAUAU-AUUCUCCUC-------UUGCAGGCACUUUCACAGCAUGGAAGUGUUUGCCACCUUUGACAUAUUCGAUGCAGAGGGUCGCAAAGUUGCCCAGGGCCACA .....(((.........-.........-------..(((((((((((((.....)))))))))))))..((((((.(((.....)))))))))))).....((.(((...))).)). ( -31.30) >DroSec_CAF1 21834 111 - 1 CUUACACCAAACCAUAUCUUUCACAAUUC------UCUAGGCAUUUUCACAGCAUGGAGGUGUUUGCCACCUUUGAUAUCUUCGAUUUAAGAGGACACAAAGUGGCUCAGGGACACA ...........................((------((((((((((((((.....)))))))))))(((((.((((...(((((.......)))))..)))))))))..))))).... ( -27.40) >DroSim_CAF1 23721 107 - 1 UUUACACCAUUUC----CUAUCGAAAUUC------CUUAGGCAUUUUCACAGCAUGGAGGUGUUUGCCACCUUUGAUAUCUUCGAUUUAAGAGGACACAAAGUGGCUCAGGGACACA ........(((((----.....)))))((------((((((((((((((.....)))))))))))(((((.((((...(((((.......)))))..)))))))))..))))).... ( -28.20) >DroAna_CAF1 8376 108 - 1 CUUACUCUACCUAA---UAUUCCAUCUUC------CCCAGGCACUAUCACAGCAUGGAGGUGUUUGCCACUUUUGACAUCUUCGACCUGAGUGGCCGGAAAGUGGCCCAGGGCCACA ..............---..((((......------...(((((((.(((.....))).)))))))(((((((((((.....))))...))))))).)))).((((((...)))))). ( -32.70) >DroPer_CAF1 24207 117 - 1 CGUUGAACACUCAAGAGCACUCUCAUUUCUCUCUUACCAGGCACUUUCACAGCAUGGAAGUGUUUGCCACCUUUGACAUCUUCGAUUUGAGCGGUCGGAAGGUCGCUCAGGGCCACA .((..((.....(((((.............)))))..((((((((((((.....)))))))))))).....))..))........((((((((..(....)..))))))))...... ( -36.12) >consensus CUUACACCACACCAUAUCAUUCACAAUUC______UCCAGGCACUUUCACAGCAUGGAGGUGUUUGCCACCUUUGACAUCUUCGAUUUAAGAGGACGCAAAGUGGCUCAGGGACACA ...................................((((((((((((((.....)))))))))))(((((.((((....((((.......))))...)))))))))...)))..... (-17.11 = -17.75 + 0.64)

| Location | 17,297,802 – 17,297,906 |
|---|---|
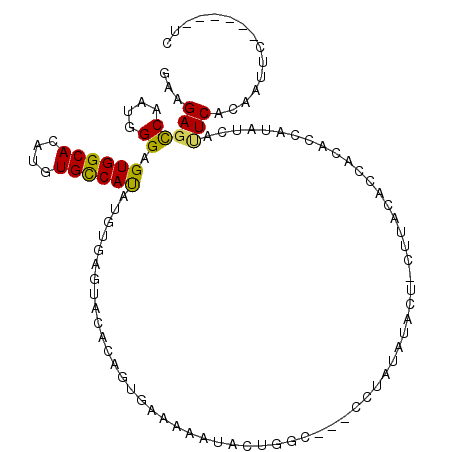
| Length | 104 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 60.52 |
| Mean single sequence MFE | -23.59 |
| Consensus MFE | -9.29 |
| Energy contribution | -8.77 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17297802 104 - 20766785 GAAGAGCCAAUGGGAGUGGCACAUGUGCCAUAUGUGAGUACACAGUGAAAAAUAGUGGCAUACCUAUAAACUAUUUACACUACACCAUAUCAUUCACAAUUC------UC (((((....((((..((((((....))))))..(((....)))((((..(((((((.............))))))).))))...)))).)).))).......------.. ( -22.42) >DroGri_CAF1 382 94 - 1 GAAGAGCCAGUGGGAGUGGCAUAUGUGUCAUAUGUAAGCAUA--AUGAUUAGUUA-GC-----CCAACUUGUGCUUAAGAUAUAAUAUAU-AUUCUCCUC-------UUG ...........(((((..(.((((((((.((((.((((((((--......((((.-..-----..))))))))))))..)))).))))))-)).)..)))-------)). ( -22.50) >DroSec_CAF1 21914 104 - 1 GAAGAGCCAAUGGGAGUGGCACAUGUGCCAUAUGUGAGUACACAGUGAAAAAUUCUGGCAAACCUAUAUACUACUUACACCAAACCAUAUCUUUCACAAUUC------UC (((((....((((..((((((....)))))).((((((((......((.....)).((....)).......)))))))).....)))).)))))........------.. ( -21.40) >DroSim_CAF1 23801 96 - 1 GAAGAGCCAAUGGGAGUGGCACAUGUGCCAUAUGUGAGUACACAGUGAAAAAUACUGGCAAAUCUAUAC----UUUACACCAUUUC----CUAUCGAAAUUC------CU (((........(((((((((((((((...)))))))......(((((.....)))))............----......)))))))----)........)))------.. ( -20.39) >DroAna_CAF1 8456 81 - 1 GAAGAGCCAGUGGGAGUGGCACAUGUGUCACAUGUAAGUGU-CAG-----------------CCUAAGGAC--CUUACUCUACCUAA---UAUUCCAUCUUC------CC (((((.....((((.((((((....))))))..(((((.((-(..-----------------......)))--)))))....)))).---.......)))))------.. ( -24.32) >DroPer_CAF1 24287 92 - 1 GAAGAGUCAGUGGGAGUGGCACAUGUGCCACAUGUGAGUAGAU---------UACUGUU----CU-----CUCCGUUGAACACUCAAGAGCACUCUCAUUUCUCUCUUAC .(((((..((((((((((.((((((.....)))))).......---------...((((----(.-----.......)))))........))))))))))...))))).. ( -30.50) >consensus GAAGAGCCAAUGGGAGUGGCACAUGUGCCAUAUGUGAGUACACAGUGAAAAAUACUGGC___CCUAUAUACU_CUUACACCACACCAUAUCAUUCACAAUUC______UC ...(((((....)).((((((....)))))).............................................................)))............... ( -9.29 = -8.77 + -0.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:00 2006