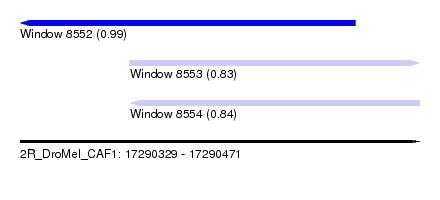
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,290,329 – 17,290,471 |
| Length | 142 |
| Max. P | 0.987030 |

| Location | 17,290,329 – 17,290,448 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.87 |
| Mean single sequence MFE | -17.16 |
| Consensus MFE | -15.11 |
| Energy contribution | -15.80 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.987030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17290329 119 - 20766785 CAAUUUUCGCACUUAAUUAUAAGCAACUCCAGCUGCCAAUUGCAGCAAAACAAACAAAAAACAAGCAAAAACAGGACACUGCACAGAAA-AAUGUGAAAAUACCAAAAAAAAAAUAAUUU ..(((((((((....................(((((.....))))).........................(((....)))........-..)))))))))................... ( -18.40) >DroSec_CAF1 14693 117 - 1 CAAUUUUCGCACUUAAUUAUAAGCAACUCCAGCUGCCAAUUGCAGCAAAACAAACAA-AAACAAGCAAAAACAGGGCACUGCGCAGAAAAAAUGUUAAAAUACCAAAUUAAAAAG--UUA ...(((((((.((((....))))........(((((.....)))))...........-......)).....(((....)))....))))).........................--... ( -15.60) >DroSim_CAF1 16402 117 - 1 CAAUUUUCGCACUUAAUUAUAAGCAACUCCAGCUGCCAAUUGCAGCAAAACAAACAA-AAACAAGCAAAAACAGGACACUGCACAGAAAAGAUGUGAAAUUACCAAAAUAAAAAU--UUU ....((((((((((........(((..(((.(((((.....)))))...........-......(......).)))...)))......))).)))))))................--... ( -17.84) >DroEre_CAF1 10447 116 - 1 CAAUUUUCGCACUUAAUUAUAAGCAAGCCCAGCUGCCAAUUGCAGCAAAGCAAACAA-AAAGCAGCAAAAACAGGACAAUGCACUGAAAAAUUGUUGAAAUACCAAAAUAAAAG---UUU ....((((((.((((....))))...((...(((((.....)))))...))......-...))...........((((((..........))))))))))..............---... ( -16.80) >consensus CAAUUUUCGCACUUAAUUAUAAGCAACUCCAGCUGCCAAUUGCAGCAAAACAAACAA_AAACAAGCAAAAACAGGACACUGCACAGAAAAAAUGUGAAAAUACCAAAAUAAAAAU__UUU ..(((((((((...........(((..(((.(((((.....)))))..................(......).)))...)))..........)))))))))................... (-15.11 = -15.80 + 0.69)

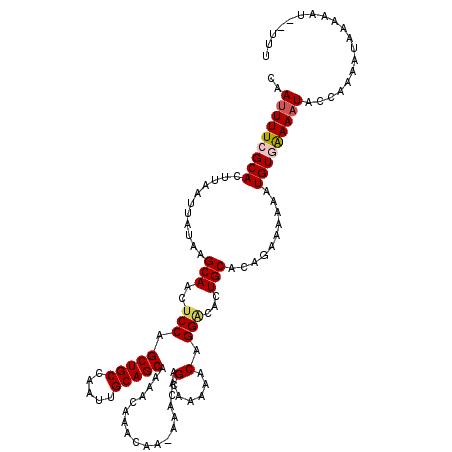
| Location | 17,290,368 – 17,290,471 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 94.63 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -27.38 |
| Energy contribution | -26.88 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17290368 103 + 20766785 AGUGUCCUGUUUUUGCUUGUUUUUUGUUUGUUUUGCUGCAAUUGGCAGCUGGAGUUGCUUAUAAUUAAGUGCGAAAAUUGGCCACAGACUCGAGUGGAGUCCC .(((.((.((((((((..............((..(((((.....)))))..))...(((((....))))))))))))).)).))).(((((.....))))).. ( -32.00) >DroSec_CAF1 14731 102 + 1 AGUGCCCUGUUUUUGCUUGUUU-UUGUUUGUUUUGCUGCAAUUGGCAGCUGGAGUUGCUUAUAAUUAAGUGCGAAAAUUGGCCACAGGUUCGAGUGGAGUCCC .(((.((.((((((((......-.......((..(((((.....)))))..))...(((((....))))))))))))).)).))).(((((.....))).)). ( -27.90) >DroSim_CAF1 16440 102 + 1 AGUGUCCUGUUUUUGCUUGUUU-UUGUUUGUUUUGCUGCAAUUGGCAGCUGGAGUUGCUUAUAAUUAAGUGCGAAAAUUGGCCACAGGUUCGAGUGGAGUCGC .(((.((.((((((((......-.......((..(((((.....)))))..))...(((((....))))))))))))).)).))).(..((.....))..).. ( -28.30) >DroEre_CAF1 10484 102 + 1 AUUGUCCUGUUUUUGCUGCUUU-UUGUUUGCUUUGCUGCAAUUGGCAGCUGGGCUUGCUUAUAAUUAAGUGCGAAAAUUGGCCACAGGUUCGAGUGGAGUCCC .....((.((((((((.((((.-..((..(((..(((((.....)))))..)))..))........)))))))))))).))((((........))))...... ( -30.70) >consensus AGUGUCCUGUUUUUGCUUGUUU_UUGUUUGUUUUGCUGCAAUUGGCAGCUGGAGUUGCUUAUAAUUAAGUGCGAAAAUUGGCCACAGGUUCGAGUGGAGUCCC .(((.((.((((((((..............((..(((((.....)))))..))...(((((....))))))))))))).)).))).(((((.....))))).. (-27.38 = -26.88 + -0.50)

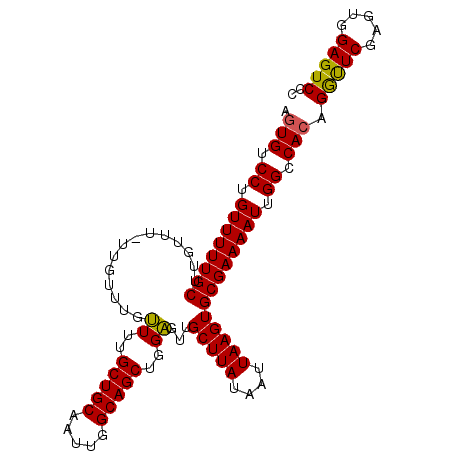
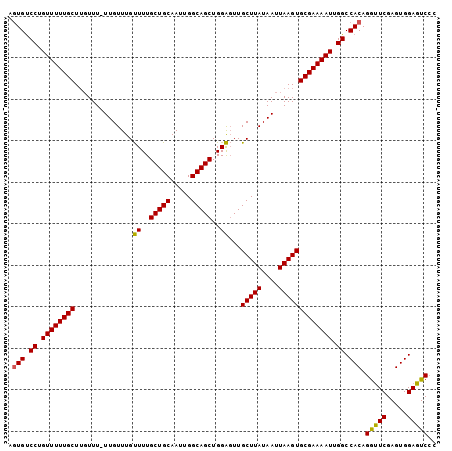
| Location | 17,290,368 – 17,290,471 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 94.63 |
| Mean single sequence MFE | -20.77 |
| Consensus MFE | -17.65 |
| Energy contribution | -17.65 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.839442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17290368 103 - 20766785 GGGACUCCACUCGAGUCUGUGGCCAAUUUUCGCACUUAAUUAUAAGCAACUCCAGCUGCCAAUUGCAGCAAAACAAACAAAAAACAAGCAAAAACAGGACACU .((((((.....))))))(((.((..((((.((.((((....))))........(((((.....)))))..................)).))))..)).))). ( -24.00) >DroSec_CAF1 14731 102 - 1 GGGACUCCACUCGAACCUGUGGCCAAUUUUCGCACUUAAUUAUAAGCAACUCCAGCUGCCAAUUGCAGCAAAACAAACAA-AAACAAGCAAAAACAGGGCACU (((......)))......(((.((..((((.((.((((....))))........(((((.....)))))...........-......)).))))..)).))). ( -20.40) >DroSim_CAF1 16440 102 - 1 GCGACUCCACUCGAACCUGUGGCCAAUUUUCGCACUUAAUUAUAAGCAACUCCAGCUGCCAAUUGCAGCAAAACAAACAA-AAACAAGCAAAAACAGGACACU .(((......)))..(((((.((........((............)).......(((((.....)))))...........-......))....)))))..... ( -18.50) >DroEre_CAF1 10484 102 - 1 GGGACUCCACUCGAACCUGUGGCCAAUUUUCGCACUUAAUUAUAAGCAAGCCCAGCUGCCAAUUGCAGCAAAGCAAACAA-AAAGCAGCAAAAACAGGACAAU (((.((((((........)))).........((............)).))))).(((((...((((......))))....-...))))).............. ( -20.20) >consensus GGGACUCCACUCGAACCUGUGGCCAAUUUUCGCACUUAAUUAUAAGCAACUCCAGCUGCCAAUUGCAGCAAAACAAACAA_AAACAAGCAAAAACAGGACACU ......((((........))))((..((((.((.((((....))))........(((((.....)))))..................)).))))..))..... (-17.65 = -17.65 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:53 2006