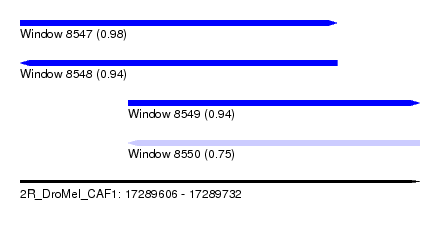
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,289,606 – 17,289,732 |
| Length | 126 |
| Max. P | 0.980429 |

| Location | 17,289,606 – 17,289,706 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.19 |
| Mean single sequence MFE | -30.25 |
| Consensus MFE | -18.21 |
| Energy contribution | -18.27 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
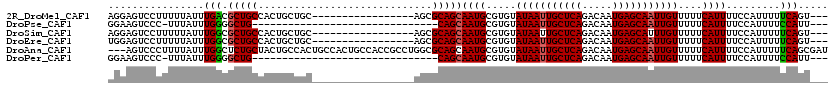
>2R_DroMel_CAF1 17289606 100 + 20766785 AGGAGUCCUUUUUAUUUGACGCUGCCACUGCUGC-----------------AGCGCAGCAAUGCGUGUAUAAUUGCUCAGACAAUGAGCAAUUGUUUUUCAUUUUCCAUUUUUCAGU--- .((((.............((((.((...((((((-----------------...))))))..))))))(((((((((((.....)))))))))))........))))..........--- ( -29.70) >DroPse_CAF1 15948 85 + 1 GGAAGUCCC-UUUAUUUGGGGCUG-------------------------------CAGCAAUGCGUGUAUAAUUGCUCAGACAAUGAGCAAUUGUUUUUCAUUUUCCAUUUUCCAUU--- (((((((((-.......))))))(-------------------------------((....)))(((.(((((((((((.....)))))))))))....)))..)))..........--- ( -30.10) >DroSim_CAF1 15680 100 + 1 AGGAGUCCUUUUUAUUUGGCGCUGCCACUGCUGC-----------------AGCGCAGCAAUGCGUGUAUAAUUGCUCAGACAAUGAGCAUUUGUUUUUCAUUUUCCAUUUUUCAGU--- .((((.............(((((((.......))-----------------)))))...((((.....((((.((((((.....)))))).))))....))))))))..........--- ( -28.10) >DroEre_CAF1 9713 100 + 1 UGGAGUCCUUUUUAUUUGGCGCUGCCACUGCUGC-----------------AGCGCAGCAAUGCGUGUAUAAUUGCUCAGACAAUGAGCAAUUGUUUUUCAUUUUCCAUUUUUCAGU--- (((((.............(((((((.......))-----------------)))))...((((.....(((((((((((.....)))))))))))....))))))))).........--- ( -31.60) >DroAna_CAF1 1158 117 + 1 ---AGUCCCUUUUAUUUGGCUCUGCUACUGCCACUGCCACUGCCACCGCCUGGCGCAGCAAUGCGUGUAUAAUUGCUCAGACAAUGAGCAAUUGUUUUUCAUUUUCCAUUUUUCAGCGAU ---.............((((...((....))....)))).......(((.(((((((....)))))..(((((((((((.....)))))))))))..................))))).. ( -31.90) >DroPer_CAF1 15853 85 + 1 GGAAGUCCC-UUUAUUUGGGGCUG-------------------------------CAGCAAUGCGUGUAUAAUUGCUCAGACAAUGAGCAAUUGUUUUUCAUUUUCCAUUUUCCAUU--- (((((((((-.......))))))(-------------------------------((....)))(((.(((((((((((.....)))))))))))....)))..)))..........--- ( -30.10) >consensus AGGAGUCCCUUUUAUUUGGCGCUGCCACUGCUGC_________________AGCGCAGCAAUGCGUGUAUAAUUGCUCAGACAAUGAGCAAUUGUUUUUCAUUUUCCAUUUUUCAGU___ ................(((.(((((.............................)))))((((.....(((((((((((.....)))))))))))....)))).........)))..... (-18.21 = -18.27 + 0.06)
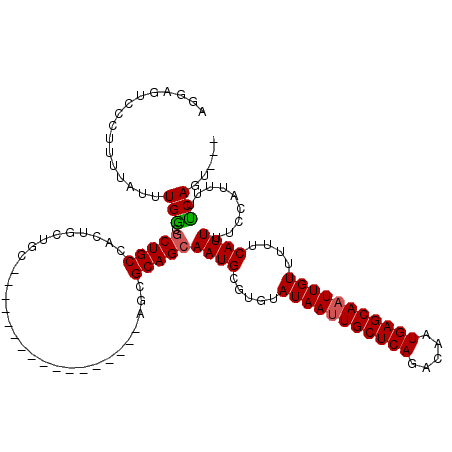
| Location | 17,289,606 – 17,289,706 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.19 |
| Mean single sequence MFE | -25.43 |
| Consensus MFE | -16.96 |
| Energy contribution | -17.24 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17289606 100 - 20766785 ---ACUGAAAAAUGGAAAAUGAAAAACAAUUGCUCAUUGUCUGAGCAAUUAUACACGCAUUGCUGCGCU-----------------GCAGCAGUGGCAGCGUCAAAUAAAAAGGACUCCU ---..........(((...........(((((((((.....)))))))))......((((((((((...-----------------)))))))).))...(((..........)))))). ( -26.60) >DroPse_CAF1 15948 85 - 1 ---AAUGGAAAAUGGAAAAUGAAAAACAAUUGCUCAUUGUCUGAGCAAUUAUACACGCAUUGCUG-------------------------------CAGCCCCAAAUAAA-GGGACUUCC ---...((((..((.(.((((......(((((((((.....))))))))).......))))..).-------------------------------))..(((.......-)))..)))) ( -18.52) >DroSim_CAF1 15680 100 - 1 ---ACUGAAAAAUGGAAAAUGAAAAACAAAUGCUCAUUGUCUGAGCAAUUAUACACGCAUUGCUGCGCU-----------------GCAGCAGUGGCAGCGCCAAAUAAAAAGGACUCCU ---..........(((.(((((...........)))))((((.((((((.........))))))(((((-----------------((.......)))))))..........))))))). ( -24.40) >DroEre_CAF1 9713 100 - 1 ---ACUGAAAAAUGGAAAAUGAAAAACAAUUGCUCAUUGUCUGAGCAAUUAUACACGCAUUGCUGCGCU-----------------GCAGCAGUGGCAGCGCCAAAUAAAAAGGACUCCA ---.........((((.((((......(((((((((.....))))))))).......))))...(((((-----------------((.......)))))))..............)))) ( -27.92) >DroAna_CAF1 1158 117 - 1 AUCGCUGAAAAAUGGAAAAUGAAAAACAAUUGCUCAUUGUCUGAGCAAUUAUACACGCAUUGCUGCGCCAGGCGGUGGCAGUGGCAGUGGCAGUAGCAGAGCCAAAUAAAAGGGACU--- .((.((......(((............(((((((((.....)))))))))......(((((((..(((((.((....))..)))).)..))))).))....)))......)).))..--- ( -36.60) >DroPer_CAF1 15853 85 - 1 ---AAUGGAAAAUGGAAAAUGAAAAACAAUUGCUCAUUGUCUGAGCAAUUAUACACGCAUUGCUG-------------------------------CAGCCCCAAAUAAA-GGGACUUCC ---...((((..((.(.((((......(((((((((.....))))))))).......))))..).-------------------------------))..(((.......-)))..)))) ( -18.52) >consensus ___ACUGAAAAAUGGAAAAUGAAAAACAAUUGCUCAUUGUCUGAGCAAUUAUACACGCAUUGCUGCGCU_________________GCAGCAGUGGCAGCGCCAAAUAAAAAGGACUCCC ............(((..((((......(((((((((.....))))))))).......))))(((((.............................))))).)))................ (-16.96 = -17.24 + 0.28)

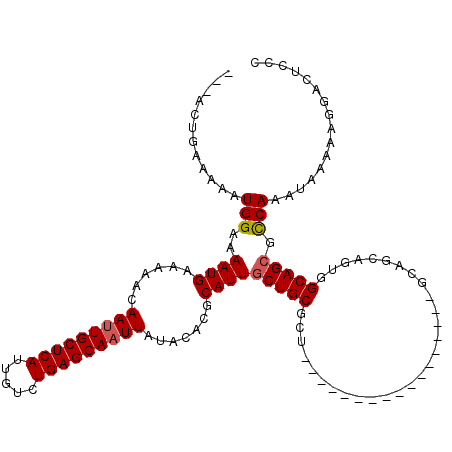

| Location | 17,289,640 – 17,289,732 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.70 |
| Mean single sequence MFE | -22.21 |
| Consensus MFE | -17.62 |
| Energy contribution | -17.79 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17289640 92 + 20766785 -----------AGCGCAGCAAUGCGUGUAUAAUUGCUCAGACAAUGAGCAAUUGUUUUUCAUUUUCCAUUUUUCAGU-----GUU---------U---UUUUCGUUGUUUGCUUGUUGAU -----------((((.(((((((.(((.(((((((((((.....)))))))))))....)))....(((......))-----)..---------.---....)))))))))))....... ( -22.10) >DroPse_CAF1 15971 94 + 1 ---------------CAGCAAUGCGUGUAUAAUUGCUCAGACAAUGAGCAAUUGUUUUUCAUUUUCCAUUUUCCAUU-----U------GCCAUUUUUCCCCCCUUGUUUGCUUGUUGAU ---------------((((((.(((((.(((((((((((.....)))))))))))....)))....((.........-----)------)....................)))))))).. ( -19.30) >DroSim_CAF1 15714 92 + 1 -----------AGCGCAGCAAUGCGUGUAUAAUUGCUCAGACAAUGAGCAUUUGUUUUUCAUUUUCCAUUUUUCAGU-----GUU---------U---UUUUCGUUGUUUGCUUGUUGAU -----------((((.(((((((.(((.((((.((((((.....)))))).))))....)))....(((......))-----)..---------.---....)))))))))))....... ( -18.90) >DroEre_CAF1 9747 92 + 1 -----------AGCGCAGCAAUGCGUGUAUAAUUGCUCAGACAAUGAGCAAUUGUUUUUCAUUUUCCAUUUUUCAGU-----GUU---------U---UUUUCGUUGUUUGCUUGUUGAU -----------((((.(((((((.(((.(((((((((((.....)))))))))))....)))....(((......))-----)..---------.---....)))))))))))....... ( -22.10) >DroAna_CAF1 1195 120 + 1 UGCCACCGCCUGGCGCAGCAAUGCGUGUAUAAUUGCUCAGACAAUGAGCAAUUGUUUUUCAUUUUCCAUUUUUCAGCGAUGUGUUUUUUGUGUUUUUUGUUUCGUUGUUUGCUUGUUGAU .((((.....)))).((((((.(((((.(((((((((((.....)))))))))))....)))...........((((((......................))))))...)))))))).. ( -31.55) >DroPer_CAF1 15876 94 + 1 ---------------CAGCAAUGCGUGUAUAAUUGCUCAGACAAUGAGCAAUUGUUUUUCAUUUUCCAUUUUCCAUU-----U------GCCAUUUUUCCCCCGUUGUUUGCUUGUUGAU ---------------((((((.(((((.(((((((((((.....)))))))))))....)))....((.........-----)------)....................)))))))).. ( -19.30) >consensus ___________AGCGCAGCAAUGCGUGUAUAAUUGCUCAGACAAUGAGCAAUUGUUUUUCAUUUUCCAUUUUUCAGU_____GUU_________U___UUUUCGUUGUUUGCUUGUUGAU ...............((((((.(((((.(((((((((((.....)))))))))))....)))....((.....................................))...)))))))).. (-17.62 = -17.79 + 0.17)

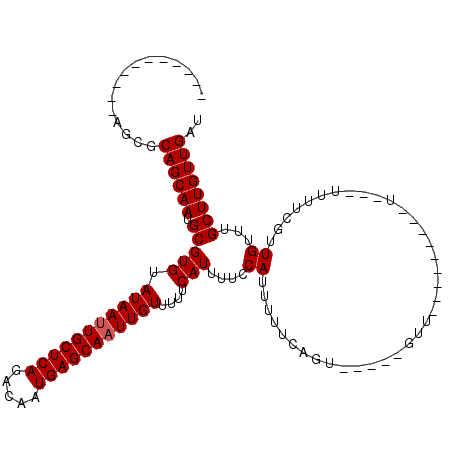
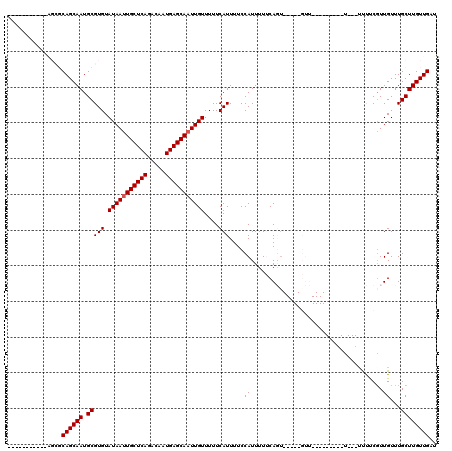
| Location | 17,289,640 – 17,289,732 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.70 |
| Mean single sequence MFE | -15.77 |
| Consensus MFE | -12.60 |
| Energy contribution | -12.77 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17289640 92 - 20766785 AUCAACAAGCAAACAACGAAAA---A---------AAC-----ACUGAAAAAUGGAAAAUGAAAAACAAUUGCUCAUUGUCUGAGCAAUUAUACACGCAUUGCUGCGCU----------- .......(((............---.---------...-----........................(((((((((.....)))))))))......(((....))))))----------- ( -15.10) >DroPse_CAF1 15971 94 - 1 AUCAACAAGCAAACAAGGGGGGAAAAAUGGC------A-----AAUGGAAAAUGGAAAAUGAAAAACAAUUGCUCAUUGUCUGAGCAAUUAUACACGCAUUGCUG--------------- .......(((((.(......)........((------.-----..(....)................(((((((((.....)))))))))......)).))))).--------------- ( -13.40) >DroSim_CAF1 15714 92 - 1 AUCAACAAGCAAACAACGAAAA---A---------AAC-----ACUGAAAAAUGGAAAAUGAAAAACAAAUGCUCAUUGUCUGAGCAAUUAUACACGCAUUGCUGCGCU----------- .......(((............---.---------...-----...........................((((((.....)))))).........(((....))))))----------- ( -12.30) >DroEre_CAF1 9747 92 - 1 AUCAACAAGCAAACAACGAAAA---A---------AAC-----ACUGAAAAAUGGAAAAUGAAAAACAAUUGCUCAUUGUCUGAGCAAUUAUACACGCAUUGCUGCGCU----------- .......(((............---.---------...-----........................(((((((((.....)))))))))......(((....))))))----------- ( -15.10) >DroAna_CAF1 1195 120 - 1 AUCAACAAGCAAACAACGAAACAAAAAACACAAAAAACACAUCGCUGAAAAAUGGAAAAUGAAAAACAAUUGCUCAUUGUCUGAGCAAUUAUACACGCAUUGCUGCGCCAGGCGGUGGCA .......................................(((((((......(((............(((((((((.....))))))))).....((((....))))))))))))))... ( -25.10) >DroPer_CAF1 15876 94 - 1 AUCAACAAGCAAACAACGGGGGAAAAAUGGC------A-----AAUGGAAAAUGGAAAAUGAAAAACAAUUGCUCAUUGUCUGAGCAAUUAUACACGCAUUGCUG--------------- .......(((((....((...(........)------.-----..))....................(((((((((.....))))))))).........))))).--------------- ( -13.60) >consensus AUCAACAAGCAAACAACGAAAA___A_________AAC_____ACUGAAAAAUGGAAAAUGAAAAACAAUUGCUCAUUGUCUGAGCAAUUAUACACGCAUUGCUGCGCU___________ .......(((((........................................(....).........(((((((((.....))))))))).........)))))................ (-12.60 = -12.77 + 0.17)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:49 2006