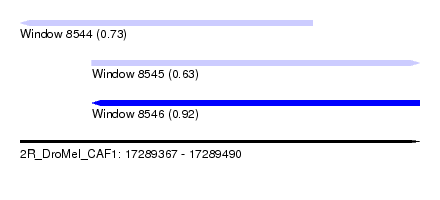
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,289,367 – 17,289,490 |
| Length | 123 |
| Max. P | 0.922735 |

| Location | 17,289,367 – 17,289,457 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 94.18 |
| Mean single sequence MFE | -20.50 |
| Consensus MFE | -18.60 |
| Energy contribution | -18.93 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.727059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
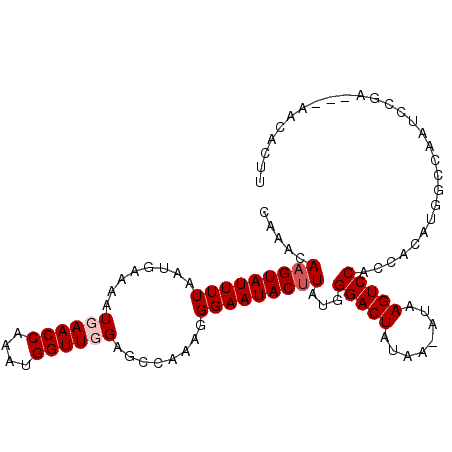
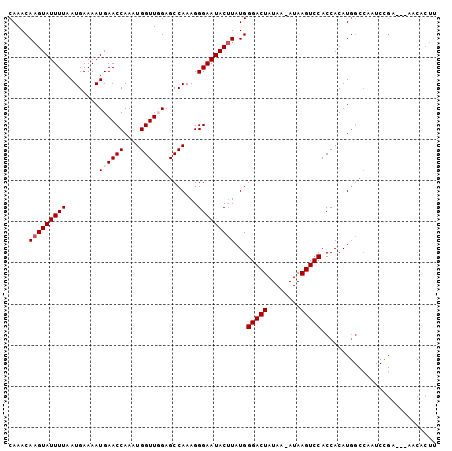
>2R_DroMel_CAF1 17289367 90 - 20766785 GUUGGAGCCAAAGGGAAUACUUAUGGGACUAUAAAAUAAGUCCACCACAUGGCCAAUCUGA---AACACUUUAUCCAACGGCACACUUAAGCA ((((((((((...((....(....)(((((........))))).))...)))).....(((---(....)))))))))).((........)). ( -21.20) >DroSec_CAF1 13731 89 - 1 GUUUGAGCCAAAGGGAAUACUUAUGGGACUAUAA-AUAAGUCCACCACAUGGCCAAUCCGA---AACACUUUAUCCAACGGCGCAAUUAAGCA (((((((((....(((.........(((((....-...))))).((....)).........---.........)))...)))....)))))). ( -17.40) >DroSim_CAF1 15439 92 - 1 GUUGGAGCCAAAGGGAAUACUUAUGGGACUAUAA-AUAAGUCCACCACAUGGCCAAUCCGAAGAAACACUUUAUCCAACGGCACAAUUAAGCA ((((((((((...((....(....)(((((....-...))))).))...))))......((((.....)))).)))))).((........)). ( -22.90) >consensus GUUGGAGCCAAAGGGAAUACUUAUGGGACUAUAA_AUAAGUCCACCACAUGGCCAAUCCGA___AACACUUUAUCCAACGGCACAAUUAAGCA ((((((((((...((....(....)(((((........))))).))...))))......(......)......)))))).((........)). (-18.60 = -18.93 + 0.33)

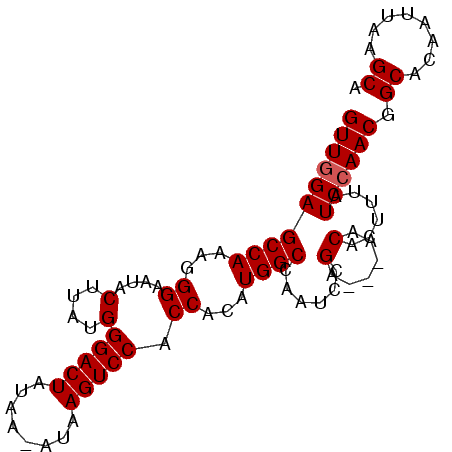
| Location | 17,289,389 – 17,289,490 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 87.25 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -14.76 |
| Energy contribution | -14.82 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17289389 101 + 20766785 AAGUGUU---UCAGAUUGGCCAUGUGGUGGACUUAUUUUAUAGUCCCAUAAGUAUUCCCUUUGGCUCCAACCAUUUGGUUCAUUUUCAUUAAAAUACUUGUUUG (((((((---(.((((..((((.((((((((((........)))))...............((....))))))).))))..))))......))))))))..... ( -22.50) >DroSec_CAF1 13753 100 + 1 AAGUGUU---UCGGAUUGGCCAUGUGGUGGACUUAU-UUAUAGUCCCAUAAGUAUUCCCUUUGGCUCAAACCAUUUGGUUCAUUUUCAUUAAAAUACUUGUUUG (((((((---(..(.(((((((...((.(((..(((-((((......))))))).))))).)))).)))(((....))).........)..))))))))..... ( -22.90) >DroSim_CAF1 15461 103 + 1 AAGUGUUUCUUCGGAUUGGCCAUGUGGUGGACUUAU-UUAUAGUCCCAUAAGUAUUCCCUUUGGCUCCAACCAUUUGGUUCAUUUUCAUUAAAAUACUUGUUUG ((((((((....(((...((((...((.(((..(((-((((......))))))).))))).)))))))((((....))))...........))))))))..... ( -23.50) >DroEre_CAF1 9508 82 + 1 ---------------------AUGUGGUGGACUUAU-UUAUAGUCCCAAAGGUAUUCCCUUUGGCUCAAACCAUUUGGUUCAUUUUCAUUAAAAUACUUGUUUG ---------------------..((((((((((...-....)))))((((((.....))))))......))))).............................. ( -16.70) >consensus AAGUGUU___UCGGAUUGGCCAUGUGGUGGACUUAU_UUAUAGUCCCAUAAGUAUUCCCUUUGGCUCAAACCAUUUGGUUCAUUUUCAUUAAAAUACUUGUUUG ..........................(.(((((........))))))(((((((((.....(((....((((....)))).....)))....)))))))))... (-14.76 = -14.82 + 0.06)


| Location | 17,289,389 – 17,289,490 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 87.25 |
| Mean single sequence MFE | -19.85 |
| Consensus MFE | -15.54 |
| Energy contribution | -16.29 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17289389 101 - 20766785 CAAACAAGUAUUUUAAUGAAAAUGAACCAAAUGGUUGGAGCCAAAGGGAAUACUUAUGGGACUAUAAAAUAAGUCCACCACAUGGCCAAUCUGA---AACACUU .....((((.(((((.......(.((((....)))).).((((...((....(....)(((((........))))).))...)))).....)))---)).)))) ( -20.10) >DroSec_CAF1 13753 100 - 1 CAAACAAGUAUUUUAAUGAAAAUGAACCAAAUGGUUUGAGCCAAAGGGAAUACUUAUGGGACUAUAA-AUAAGUCCACCACAUGGCCAAUCCGA---AACACUU .....((((.((((.........(((((....)))))(.((((...((....(....)(((((....-...))))).))...))))).....))---)).)))) ( -19.80) >DroSim_CAF1 15461 103 - 1 CAAACAAGUAUUUUAAUGAAAAUGAACCAAAUGGUUGGAGCCAAAGGGAAUACUUAUGGGACUAUAA-AUAAGUCCACCACAUGGCCAAUCCGAAGAAACACUU .....((((.((((..........((((....))))(((((((...((....(....)(((((....-...))))).))...))))...)))...)))).)))) ( -21.20) >DroEre_CAF1 9508 82 - 1 CAAACAAGUAUUUUAAUGAAAAUGAACCAAAUGGUUUGAGCCAAAGGGAAUACCUUUGGGACUAUAA-AUAAGUCCACCACAU--------------------- .......................(((((....)))))....((((((.....))))))(((((....-...))))).......--------------------- ( -18.30) >consensus CAAACAAGUAUUUUAAUGAAAAUGAACCAAAUGGUUGGAGCCAAAGGGAAUACUUAUGGGACUAUAA_AUAAGUCCACCACAUGGCCAAUCCGA___AACACUU .....(((((((((........((((((....))))))........)))))))))...(((((........)))))............................ (-15.54 = -16.29 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:46 2006