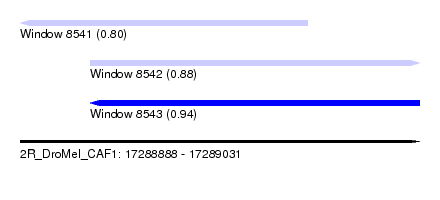
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,288,888 – 17,289,031 |
| Length | 143 |
| Max. P | 0.936231 |

| Location | 17,288,888 – 17,288,991 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -26.09 |
| Consensus MFE | -16.09 |
| Energy contribution | -17.96 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
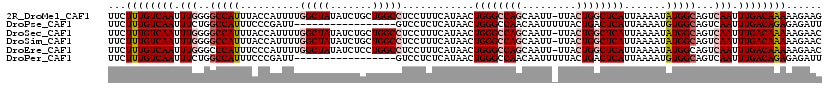
>2R_DroMel_CAF1 17288888 103 - 20766785 CUGCUGGCCUCCUUUCAUAACUGGGCCAGCAAUU-UUACUGGCUCAUUAAAAUAUGGCAGUCAAUUUGACAAAAAGA-AGCCC--------GGG-------UAAUAACAGCAUCCUCACA .(((((((((..((....))..)))))))))...-((((((((((((......)))...(((.....))).......-)))).--------.))-------)))................ ( -26.40) >DroPse_CAF1 15299 96 - 1 --------GUCCUCUCAUAACUGGGCCAACAAUUUUUACUGACUCAUUAAAAUGUGGCAGUCAAUUUGACAGAGAGA-UUCCCUUAGGGAUGGG-------UGGCACGAGUA-------- --------((((((((........((((...(((((............))))).)))).(((.....))).))))).-..(((........)))-------.))).......-------- ( -22.50) >DroSec_CAF1 13258 103 - 1 CUGCUGGCCUCCUUUCAUAACUGGGCCAGCAAUU-UUACUGGCUCAUUAAAAUAUGGCAGUCAAUUUGACAAAAAGA-ACCCC--------GGG-------UAAUAACAGCAUCAUCGCA .(((((((((..((....))..)))))))))...-(((((.((.(((......))))).(((.....))).......-.....--------.))-------))).....((......)). ( -26.20) >DroSim_CAF1 14966 103 - 1 CUGCUGGCCUCCUUUCAUAACUGGGCCAGCAAUU-UUACUGGCUCAUUAAAAUAUGGCAGUCAAUUUGACAAAAAGA-ACCCC--------GGG-------UAAUAACAGCACCCUCGCA .(((((((((..((....))..)))))))))...-......((.(((......))))).(((.....))).......-.....--------(((-------(.........))))..... ( -26.20) >DroEre_CAF1 9009 103 - 1 CUCCUGGCCUCCUUUCAUAACUGGGCCAGCAAUU-UUACUGGCUCAUUAAAAUAUGGCAGUCAAUUUGACAAAAAGA-ACCCC--------GGG-------UAAUAACAGCAUCCUCCCA ..(((((((........(((.((((((((.....-...)))))))))))......))).(((.....))).......-....)--------)))-------................... ( -23.34) >DroAna_CAF1 739 112 - 1 UUGCUGGCCUCCUUUCAUAACUGGGCCGGCAAUUUUUACUGGCUCAUUAAAAUGUGGCAGUCAAUUUGACAAAAUGGAACCCU--------CUGUCCUGCCAAAAAGCCGCAUCCCCAUA ((((((((((..((....))..))))))))))........(((((((....)))((((((((.....))).....(((.....--------...))))))))...))))........... ( -31.90) >consensus CUGCUGGCCUCCUUUCAUAACUGGGCCAGCAAUU_UUACUGGCUCAUUAAAAUAUGGCAGUCAAUUUGACAAAAAGA_ACCCC________GGG_______UAAUAACAGCAUCCUCACA .((((((((........(((.((((((((.........)))))))))))......))).(((.....))).....................................)))))........ (-16.09 = -17.96 + 1.86)

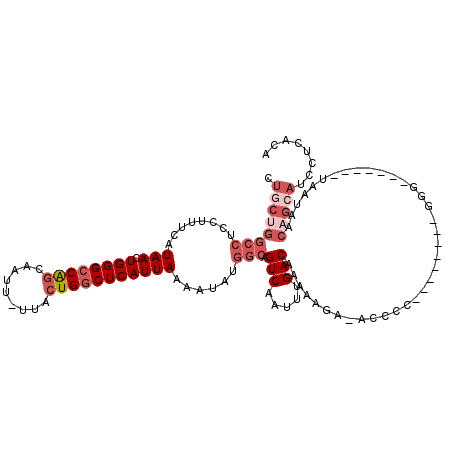
| Location | 17,288,913 – 17,289,031 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.79 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -24.67 |
| Energy contribution | -25.45 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17288913 118 + 20766785 CUUCUUUUUGUCAAAUUGACUGCCAUAUUUUAAUGAGCCAGUAA-AAUUGCUGGCCCAGUUAUGAAAGGAGGCCAGCAGAUAUAGCCAAAAUGGUAAAUGGCCCCAAAUUGACAAAGAA ....((((((((((..((((((.(((......))).(((((((.-...))))))).)))))).....((.(((((.........(((.....)))...)))))))...)))))))))). ( -37.50) >DroPse_CAF1 15324 102 + 1 AAUCUCUCUGUCAAAUUGACUGCCACAUUUUAAUGAGUCAGUAAAAAUUGUUGGCCCAGUUAUGAGAGGAC-----------------AAUCGGGAAAUGGCCAGAAAUUGACAAAGAA ....(((.(((((((((((((..............)))))))........((((((...((.(((......-----------------..))).))...))))))...)))))).))). ( -24.54) >DroSec_CAF1 13283 118 + 1 GUUCUUUUUGUCAAAUUGACUGCCAUAUUUUAAUGAGCCAGUAA-AAUUGCUGGCCCAGUUAUGAAAGGAGGCCAGCAGAUAUAGCCAAAAUGGUAAAUGGCCCCAAAUUGACAAAGAA ....((((((((((..((((((.(((......))).(((((((.-...))))))).)))))).....((.(((((.........(((.....)))...)))))))...)))))))))). ( -37.50) >DroSim_CAF1 14991 118 + 1 GUUCUUUUUGUCAAAUUGACUGCCAUAUUUUAAUGAGCCAGUAA-AAUUGCUGGCCCAGUUAUGAAAGGAGGCCAGCAGAUAUAGCCAAAAUGGUAAAUGGCCCCAAAUUGACAAAGAA ....((((((((((..((((((.(((......))).(((((((.-...))))))).)))))).....((.(((((.........(((.....)))...)))))))...)))))))))). ( -37.50) >DroEre_CAF1 9034 118 + 1 GUUCUUUUUGUCAAAUUGACUGCCAUAUUUUAAUGAGCCAGUAA-AAUUGCUGGCCCAGUUAUGAAAGGAGGCCAGGAGAUAUAGCCAAAAUGGGAAAUGGGCCCAAAUUGACAAAGAA ....(((((((((((((..(((((...(((((.((.(((((((.-...))))))).))....)))))...)).)))..)))..........((((.......))))..)))))))))). ( -35.20) >DroPer_CAF1 15228 102 + 1 AAUCUCUCUGUCAAAUUGACUGCCACAUUUUAAUGAGUCAGUAAAAAUUGUUGGCCCAGUUAUGAGAGGAC-----------------AAUCGGGAAAUGGCCAGAAAUUGACAAAGAA ....(((.(((((((((((((..............)))))))........((((((...((.(((......-----------------..))).))...))))))...)))))).))). ( -24.54) >consensus GUUCUUUUUGUCAAAUUGACUGCCAUAUUUUAAUGAGCCAGUAA_AAUUGCUGGCCCAGUUAUGAAAGGAGGCCAGCAGAUAUAGCCAAAAUGGGAAAUGGCCCCAAAUUGACAAAGAA ....((((((((((.(((...(((((.(((((.((.(((((((.....))))))).))....)))))...(((...........)))..........)))))..))).)))))))))). (-24.67 = -25.45 + 0.78)

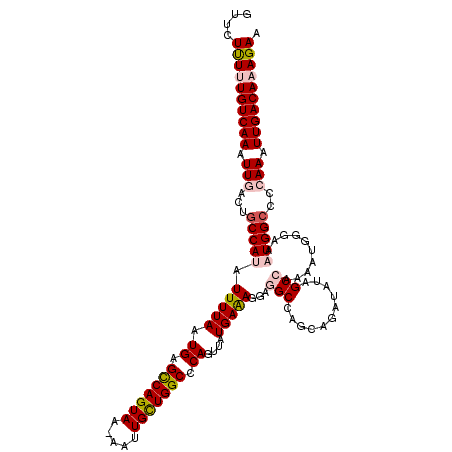

| Location | 17,288,913 – 17,289,031 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 84.79 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -24.34 |
| Energy contribution | -25.73 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.07 |
| Mean z-score | -4.04 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
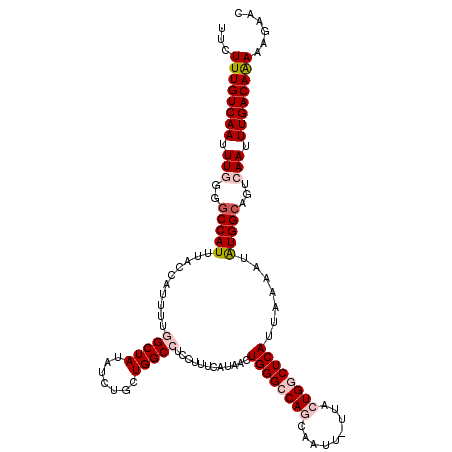
>2R_DroMel_CAF1 17288913 118 - 20766785 UUCUUUGUCAAUUUGGGGCCAUUUACCAUUUUGGCUAUAUCUGCUGGCCUCCUUUCAUAACUGGGCCAGCAAUU-UUACUGGCUCAUUAAAAUAUGGCAGUCAAUUUGACAAAAAGAAG (((((((((((.((((.(((((..........(((((((..(((((((((..((....))..)))))))))...-.)).))))).........)))))..)))).))))))..))))). ( -38.91) >DroPse_CAF1 15324 102 - 1 UUCUUUGUCAAUUUCUGGCCAUUUCCCGAUU-----------------GUCCUCUCAUAACUGGGCCAACAAUUUUUACUGACUCAUUAAAAUGUGGCAGUCAAUUUGACAGAGAGAUU (((((((((((....(((((.......((..-----------------......)).......)))))...........((((((((......)))..)))))..)))))))))))... ( -25.34) >DroSec_CAF1 13283 118 - 1 UUCUUUGUCAAUUUGGGGCCAUUUACCAUUUUGGCUAUAUCUGCUGGCCUCCUUUCAUAACUGGGCCAGCAAUU-UUACUGGCUCAUUAAAAUAUGGCAGUCAAUUUGACAAAAAGAAC (((((((((((.((((.(((((..........(((((((..(((((((((..((....))..)))))))))...-.)).))))).........)))))..)))).))))))..))))). ( -38.61) >DroSim_CAF1 14991 118 - 1 UUCUUUGUCAAUUUGGGGCCAUUUACCAUUUUGGCUAUAUCUGCUGGCCUCCUUUCAUAACUGGGCCAGCAAUU-UUACUGGCUCAUUAAAAUAUGGCAGUCAAUUUGACAAAAAGAAC (((((((((((.((((.(((((..........(((((((..(((((((((..((....))..)))))))))...-.)).))))).........)))))..)))).))))))..))))). ( -38.61) >DroEre_CAF1 9034 118 - 1 UUCUUUGUCAAUUUGGGCCCAUUUCCCAUUUUGGCUAUAUCUCCUGGCCUCCUUUCAUAACUGGGCCAGCAAUU-UUACUGGCUCAUUAAAAUAUGGCAGUCAAUUUGACAAAAAGAAC (((((((((((.(((.(((.............(((((.......)))))............((((((((.....-...)))))))).........)))...))).))))))..))))). ( -33.00) >DroPer_CAF1 15228 102 - 1 UUCUUUGUCAAUUUCUGGCCAUUUCCCGAUU-----------------GUCCUCUCAUAACUGGGCCAACAAUUUUUACUGACUCAUUAAAAUGUGGCAGUCAAUUUGACAGAGAGAUU (((((((((((....(((((.......((..-----------------......)).......)))))...........((((((((......)))..)))))..)))))))))))... ( -25.34) >consensus UUCUUUGUCAAUUUGGGGCCAUUUACCAUUUUGGCUAUAUCUGCUGGCCUCCUUUCAUAACUGGGCCAGCAAUU_UUACUGGCUCAUUAAAAUAUGGCAGUCAAUUUGACAAAAAGAAC ...((((((((.(((..(((((..........(((((.......)))))............((((((((.........)))))))).......)))))...))).))))))))...... (-24.34 = -25.73 + 1.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:43 2006