
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,284,064 – 17,284,173 |
| Length | 109 |
| Max. P | 0.878762 |

| Location | 17,284,064 – 17,284,173 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.59 |
| Mean single sequence MFE | -39.55 |
| Consensus MFE | -32.90 |
| Energy contribution | -32.32 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.878762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
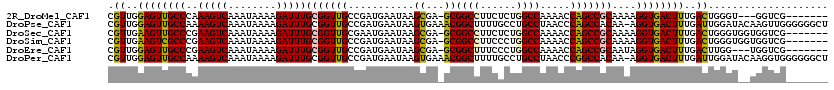
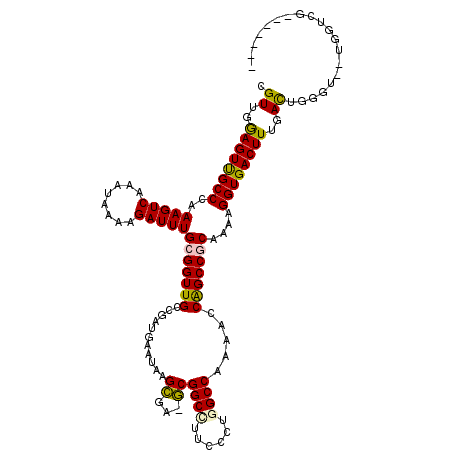
>2R_DroMel_CAF1 17284064 109 - 20766785 CGUUGGAGUUGCCCAAAGUCAAAUAAAAGAUUUGCGGUUGCCGAUGAAUAAGCGA-GCGGCCUUCUCUGGCCAAAACCAGCCGCAAAAGGUGACUUUGACUGGGU---GGUCG------- .....((.(..((((..((((((.......(((((((((((((.........)).-))((((......))))......))))))))).(....))))))))))).---.))).------- ( -39.40) >DroPse_CAF1 10364 119 - 1 CGUUGGAGUUGCCAAAAGUCAAAUAAAAGAUUUGCGGUUGCCGAUGAAUAAGUGAAACGGCUUUUGCCUGCCUAACCCAGCCACAA-AGGUGACUUUGAUUGGAUACAAGUUGGGGGGCU ((((((....(((..(((((........)))))..)))..))))))............(((....))).((((..((((((..(((-((....))))).(((....))))))))))))). ( -41.00) >DroSec_CAF1 7195 112 - 1 CGUUGAAGUUGCCCGAAGUCAAAUAAAAGAUUUGCGGUUGCGAAUGAAUAAGCGA-GCGGCCUUCUCUGGCCAAAACCAGCCGCAAAAGGUGACUUUGACUGGGUGGUGGUCG------- ....((.(..((((..(((((((.......((((((((((.(.........(...-.)((((......))))....))))))))))).(....))))))))))))..)..)).------- ( -39.20) >DroSim_CAF1 9187 112 - 1 CGUUGAAGUCGCCCGAAGUCAAAUAAAAGAUUUGCGGUUGCCGAUGAAUAAGCGA-GCGGCCUUCCCUGGCCAAAACCAGCCGCAAAAGGUGACUUUGACUGGGUGGUGGUCG------- ....((..((((((..(((((((.......(((((((((((((.........)).-))((((......))))......))))))))).(....))))))))))))))...)).------- ( -40.70) >DroEre_CAF1 3669 109 - 1 CGUUGGAGUUGCCCGAAGUCAAAUAAAAGAUUUGCGGUUGCCGAUGAAUAAGCGA-GCGGCUUUCCCUGGCCAAAACCAGCCGCAAUAGGUGACUUUGACUUGG---UGGUCG------- .((..((((..((..(((((........)))))((((((((((.........)).-))((((......))))......))))))....))..))))..))....---......------- ( -35.50) >DroPer_CAF1 10248 119 - 1 CGUUGGAGUUGCCAAAAGUCAAAUAAAAGAUUUGCGGUUGCCGAUGAAUAAGUGAAACGGCUUUUGCCUGCCUAACCCGGCCACAA-AGGUGACUUUGAUUGGAUACAAGGUGGGGGGCU ((((((....(((..(((((........)))))..)))..))))))............(((....))).(((...(((.(((.(((-((....))))).(((....)))))).)))))). ( -41.50) >consensus CGUUGGAGUUGCCCAAAGUCAAAUAAAAGAUUUGCGGUUGCCGAUGAAUAAGCGA_GCGGCCUUCCCUGGCCAAAACCAGCCGCAAAAGGUGACUUUGACUGGGU__UGGUCG_______ .((..((((((((..(((((........)))))(((((((...........((...))((((......)))).....)))))))....))))))))..)).................... (-32.90 = -32.32 + -0.58)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:38 2006