| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,282,644 – 17,282,750 |
| Length | 106 |
| Max. P | 0.527325 |

| Location | 17,282,644 – 17,282,750 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.62 |
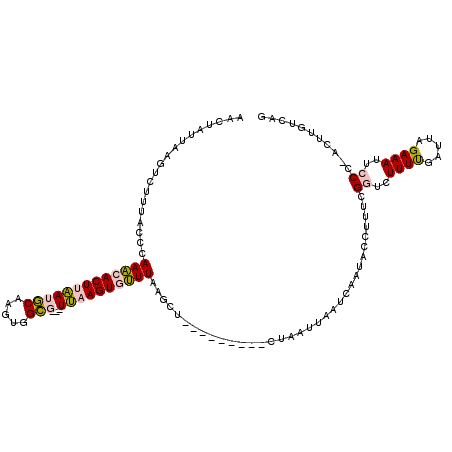
| Mean single sequence MFE | -19.97 |
| Consensus MFE | -7.80 |
| Energy contribution | -7.58 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.39 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
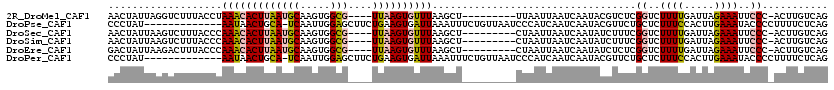
>2R_DroMel_CAF1 17282644 106 + 20766785 AACUAUUAGGUCUUUACCUAAACACUUAAUGCAAGUGGCG----UUAAGUGUUUAAGCU---------UUAAUUAAUCAAUACGUCUCGGUCUUUUGAUUAGAAAUUCCC-ACUUGUCAG ..((..(((((.......((((((((((((((.....)))----)))))))))))....---------....((((((((.((......))...))))))))........-)))))..)) ( -24.20) >DroPse_CAF1 8589 106 + 1 CCCUAU-------------AAUAACUGCA-UCAAUUGGAGCUUCUGAAGUGAUUAAAUUUCUGUUAAUCCCAUCAAUCAAUACGUUCUGCUCUUUCCACUUGAAAUACCCCUUUUCUCAG ......-------------..........-((((.(((((....(((.(.((((((.......)))))).).)))........(.....)...))))).))))................. ( -10.70) >DroSec_CAF1 5801 106 + 1 AACUAUUAAGUCUUUACCCAAACACUUAAUGCAAGUGGCG----UUAAGUGUUUAAGCU---------CUAAUUAAUCAAUAUCUUUCGGUCUUUUGAUUAGAAAUUCCC-ACUUGUCAG ..((..(((((........(((((((((((((.....)))----))))))))))....(---------((((((((.....(((....)))...))))))))).......-)))))..)) ( -24.50) >DroSim_CAF1 7782 106 + 1 AACUAUUAAGUCUUUACCCAAACACUUAAUGCAAGUGGCG----UUAAGUGUUUAAGCU---------CUAAUUAAUCAAUAUCUUUCGGUCUUUUGAUUAGAAAUUCCC-ACUUGUCAG ..((..(((((........(((((((((((((.....)))----))))))))))....(---------((((((((.....(((....)))...))))))))).......-)))))..)) ( -24.50) >DroEre_CAF1 2300 106 + 1 GACUAUUAAGACUUUACCCAAACACUUAAUGCAAGUGGCG----UUAAGUGUUUAAGCU---------CUAAUUAAUCAAUAUCUCUCGGUCUUUUGAUUAGAAAUUCCC-ACUUGUCAG .........(((.......(((((((((((((.....)))----))))))))))....(---------((((((((.....(((....)))...))))))))).......-....))).. ( -25.20) >DroPer_CAF1 8482 106 + 1 CCCUAU-------------AAUAACUGCA-UCAAUUGGAGCUUCUGAAGUGAUUAAAUUUCUGUUAAUCCCAUCAAUCAAUACGUUCUGCUCUUUCCACUUGAAAUACCCCUUUUCUCAG ......-------------..........-((((.(((((....(((.(.((((((.......)))))).).)))........(.....)...))))).))))................. ( -10.70) >consensus AACUAUUAAGUCUUUACCCAAACACUUAAUGCAAGUGGCG____UUAAGUGUUUAAGCU_________CUAAUUAAUCAAUACCUUUCGGUCUUUUGAUUAGAAAUUCCC_ACUUGUCAG ...................(((((((((((((.....)))....))))))))))..................................((..((((.....))))..))........... ( -7.80 = -7.58 + -0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:37 2006