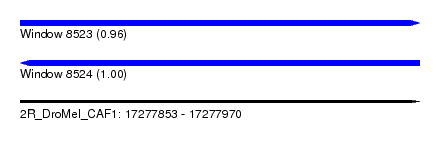
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,277,853 – 17,277,970 |
| Length | 117 |
| Max. P | 0.995610 |

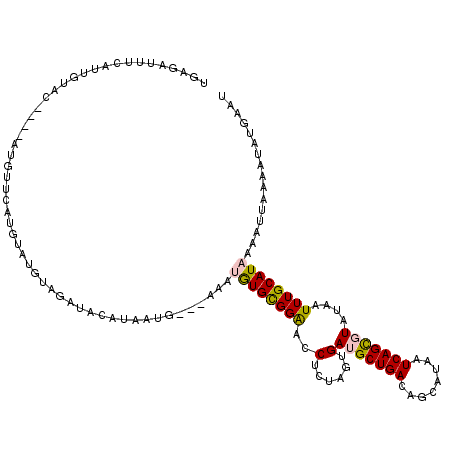
| Location | 17,277,853 – 17,277,970 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.54 |
| Mean single sequence MFE | -21.38 |
| Consensus MFE | -6.28 |
| Energy contribution | -6.68 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.29 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17277853 117 + 20766785 AAUCAUAUUUUAAUUUUAUGCAAAUUAUACGCUGAUUAUGCUGUCAGCAUCACUAAAGGUUCCGCACAUUU---CAUUAUGUAUCUACAUACAUGAACAUGUAUGUACAAUGAAAUCUCA ..................(((.........((((((......)))))).........(....)))).((((---(((..(((....((((((((....)))))))))))))))))).... ( -23.20) >DroPse_CAF1 3203 96 + 1 AUUUAUGUUUUAAUUUAAUGCAAAUUGUACACUGAUUAUGCUGUCAGCCUCGAUAGAGGCCCCACAUAUGUACACAUUUUAAUGAU----AUAUGUACAU-------------------- ...(((((.(((.(..((((.....((((((.((((......))))(((((....)))))........))))))))))..).))).----))))).....-------------------- ( -18.50) >DroSec_CAF1 876 113 + 1 ACUCAUAUUCUAAUUUUAUGCAAAUUAUACGCUGAUUAUGCUGUCAGCAUCACUGGAGGUUCCGCACAUUU---CAUUAUGUAUCUACAUACAUGAACAU----GUACAAUGAAAUCUCA ............(((((((((.........((((((......))))))......((.....))))((((((---((((((((....))))).))))).))----))...))))))).... ( -23.10) >DroSim_CAF1 2851 113 + 1 AAUCAUAUUUUAAUUUUAUGCAAAUUAUACGCUGAUUAUGCUGUCAGCAUCACUGGAGGUUCCGCACAUUU---CAUUAUGUAUCUACAUACAUGAACAU----GUACAAUGAAAUCUCA ..((((.............((.........((((((......))))))......((.....))))((((((---((((((((....))))).))))).))----))...))))....... ( -23.10) >DroPer_CAF1 3262 100 + 1 AUUCAUGUUUUAAUUUAAUGCAAAUUGUACACUGAUUAUGCUGUCAGCCUCGAUAGAGGCCCCACAUAUGUACACAUUUUAACGAUACAUAUAUGUACAU-------------------- .........................((((((.....((((.((((.(((((....))))).......(((....)))......))))))))..)))))).-------------------- ( -19.00) >consensus AAUCAUAUUUUAAUUUUAUGCAAAUUAUACGCUGAUUAUGCUGUCAGCAUCACUAGAGGUUCCGCACAUUU___CAUUAUGUAUCUACAUACAUGAACAU____GUACAAUGAAAUCUCA ..((((....((((((.....))))))...((((((......))))))............................................))))........................ ( -6.28 = -6.68 + 0.40)

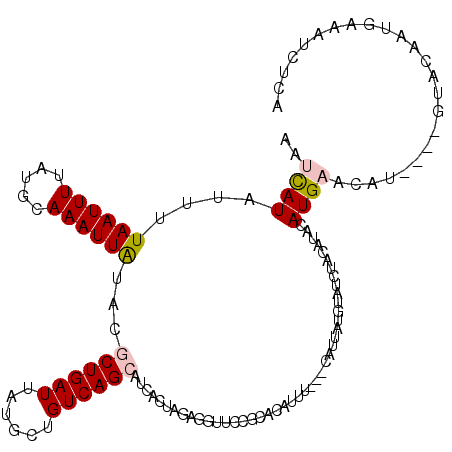
| Location | 17,277,853 – 17,277,970 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.54 |
| Mean single sequence MFE | -26.04 |
| Consensus MFE | -9.72 |
| Energy contribution | -9.56 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.37 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17277853 117 - 20766785 UGAGAUUUCAUUGUACAUACAUGUUCAUGUAUGUAGAUACAUAAUG---AAAUGUGCGGAACCUUUAGUGAUGCUGACAGCAUAAUCAGCGUAUAAUUUGCAUAAAAUUAAAAUAUGAUU ....((((((((((((((((((....))))))))......))))))---))))(((((((....(((...(((((((........))))))).))))))))))................. ( -28.20) >DroPse_CAF1 3203 96 - 1 --------------------AUGUACAUAU----AUCAUUAAAAUGUGUACAUAUGUGGGGCCUCUAUCGAGGCUGACAGCAUAAUCAGUGUACAAUUUGCAUUAAAUUAAAACAUAAAU --------------------.(((((((((----.........)))))))))(((((..((((((....))))))....)))))...((((((.....))))))................ ( -24.00) >DroSec_CAF1 876 113 - 1 UGAGAUUUCAUUGUAC----AUGUUCAUGUAUGUAGAUACAUAAUG---AAAUGUGCGGAACCUCCAGUGAUGCUGACAGCAUAAUCAGCGUAUAAUUUGCAUAAAAUUAGAAUAUGAGU ...............(----((((((((((((....))))))....---...(((((((.....))....(((((((........))))))).......)))))......)))))))... ( -26.50) >DroSim_CAF1 2851 113 - 1 UGAGAUUUCAUUGUAC----AUGUUCAUGUAUGUAGAUACAUAAUG---AAAUGUGCGGAACCUCCAGUGAUGCUGACAGCAUAAUCAGCGUAUAAUUUGCAUAAAAUUAAAAUAUGAUU .(((.((((...((((----((.(((((.(((((....))))))))---)))))))))))).))).....(((((((........))))))).((((((.....)))))).......... ( -25.30) >DroPer_CAF1 3262 100 - 1 --------------------AUGUACAUAUAUGUAUCGUUAAAAUGUGUACAUAUGUGGGGCCUCUAUCGAGGCUGACAGCAUAAUCAGUGUACAAUUUGCAUUAAAUUAAAACAUGAAU --------------------((((.((((((((((((((....))).))))))))))).((((((....))))))....))))....((((((.....))))))................ ( -26.20) >consensus UGAGAUUUCAUUGUAC____AUGUUCAUGUAUGUAGAUACAUAAUG___AAAUGUGCGGAACCUCUAGUGAUGCUGACAGCAUAAUCAGCGUAUAAUUUGCAUAAAAUUAAAAUAUGAAU ....................................................((((((((..(......)(((((((........)))))))....))))))))................ ( -9.72 = -9.56 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:25 2006