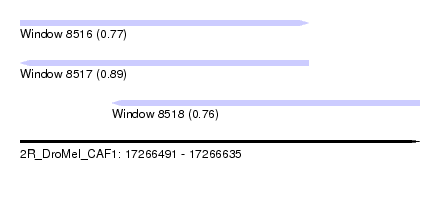
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,266,491 – 17,266,635 |
| Length | 144 |
| Max. P | 0.887863 |

| Location | 17,266,491 – 17,266,595 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.16 |
| Mean single sequence MFE | -25.63 |
| Consensus MFE | -6.22 |
| Energy contribution | -6.98 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.24 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17266491 104 + 20766785 UUU-UUUACGAUGCCCA-ACGC-UGGCCAUAAC---UUUUUCUCU-UUUUUUUUUAGCCAGCUACAAAAGCUGAAGGG------AGUGA--AAAAAAAGUGUUGCAGUGGCAUAUAACU ...-......(((((((-((((-(.........---((((((.((-(((((.......(((((.....))))))))))------)).))--))))..)))))))....)))))...... ( -23.91) >DroPse_CAF1 23399 105 + 1 UUUUUUUAUGAUCCCUAGGCGCUUGGCCAUAAU---UUUGUUCCUUUUUUUCUGCUG----------UGGCAGAAGGGGCACUCACAAAA-AAAAAAUGUUUAACAGUGGCAUAUAACU (((((((.(((......(((.....))).....---..(((((((.....(((((..----------..)))))))))))).))).))))-)))..(((((.......)))))...... ( -23.70) >DroSec_CAF1 1562 105 + 1 UUUUUUUACGAUGCCCA-ACAC-UGUCCAUAAC---UUUUUUUCGCUUUUUUUUUAGCUAGCUACAAAAGCUGAAGGG------AGUGA--AAA-AAAGUGUUGCAGUGGCAUAUAACU ..........(((((..-..((-(((.....((---(((((((((((((..((((((((.........))))))))))------)))))--)))-)))))...))))))))))...... ( -33.30) >DroEre_CAF1 1786 107 + 1 UUU-UUUACGAUGCCAA-ACGC-UGGCCAUAAUGUGUUUUUCCCU-UUUUGUUUUAGCCAGCUACAAAAGCAGCAGGG------AGUGA--AAAAAAUGUGUUGCAGUGGCAUAUAACU ...-......((((((.-..((-((((...((((...........-...))))...)))))).......((((((.(.------..(..--..)...).))))))..))))))...... ( -27.64) >DroPer_CAF1 23590 105 + 1 UUU-UUUAUGAUCCCCAGGCGCUUGGCCAUAAU---UUUGUUCUUUUUUUUUUUUUG----------UGGCAGAAGGGGCACUCAAAAAAAAAAAAAUGUUUAACAGUGGCAUAUAACU ...-.(((((.....(((....)))(((((...---.(((..(((((((((((((((----------.(((.......)).).)))))))))))))).)..)))..))))).))))).. ( -19.60) >consensus UUU_UUUACGAUGCCCA_ACGC_UGGCCAUAAU___UUUUUUCCU_UUUUUUUUUAGC_AGCUACAAAAGCAGAAGGG______AGUGA__AAAAAAUGUGUUGCAGUGGCAUAUAACU .........................(((((...................(((((((((...........)))))))))....................(.....).)))))........ ( -6.22 = -6.98 + 0.76)

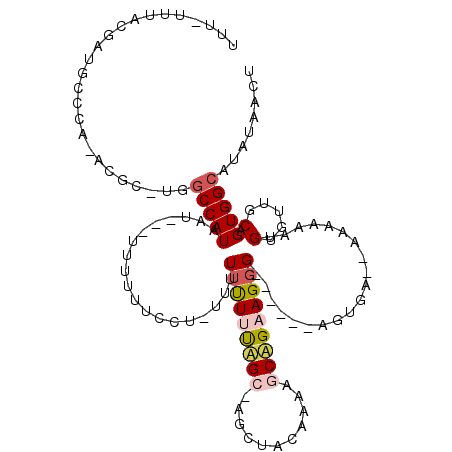

| Location | 17,266,491 – 17,266,595 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.16 |
| Mean single sequence MFE | -23.16 |
| Consensus MFE | -4.06 |
| Energy contribution | -4.38 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.18 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17266491 104 - 20766785 AGUUAUAUGCCACUGCAACACUUUUUUU--UCACU------CCCUUCAGCUUUUGUAGCUGGCUAAAAAAAAA-AGAGAAAAA---GUUAUGGCCA-GCGU-UGGGCAUCGUAAA-AAA ..(((((((((....((((.........--.....------.......((....)).((((((((........-.........---....))))))-))))-))))))).)))).-... ( -21.90) >DroPse_CAF1 23399 105 - 1 AGUUAUAUGCCACUGUUAAACAUUUUUU-UUUUGUGAGUGCCCCUUCUGCCA----------CAGCAGAAAAAAAGGAACAAA---AUUAUGGCCAAGCGCCUAGGGAUCAUAAAAAAA .........................(((-((((((((...(((.((((((..----------..)))))).............---.....(((.....)))..))).))))))))))) ( -23.70) >DroSec_CAF1 1562 105 - 1 AGUUAUAUGCCACUGCAACACUUU-UUU--UCACU------CCCUUCAGCUUUUGUAGCUAGCUAAAAAAAAAGCGAAAAAAA---GUUAUGGACA-GUGU-UGGGCAUCGUAAAAAAA ..((((((((((((((...(((((-(((--((...------......((((.....)))).(((........)))))))))))---))....).))-))..-..))))).))))..... ( -25.90) >DroEre_CAF1 1786 107 - 1 AGUUAUAUGCCACUGCAACACAUUUUUU--UCACU------CCCUGCUGCUUUUGUAGCUGGCUAAAACAAAA-AGGGAAAAACACAUUAUGGCCA-GCGU-UUGGCAUCGUAAA-AAA ..((((((((((.(((....(((.....--....(------(((((((((....)))))((.......))...-)))))..........)))....-))).-.)))))).)))).-... ( -24.61) >DroPer_CAF1 23590 105 - 1 AGUUAUAUGCCACUGUUAAACAUUUUUUUUUUUUUGAGUGCCCCUUCUGCCA----------CAAAAAAAAAAAAAGAACAAA---AUUAUGGCCAAGCGCCUGGGGAUCAUAAA-AAA ..((((((.((..((((.....((((((((((((((.(.((.......))).----------)))))))))))))).))))..---.....(((.....)))..)).)).)))).-... ( -19.70) >consensus AGUUAUAUGCCACUGCAACACAUUUUUU__UCACU______CCCUUCUGCUUUUGUAGCU_GCUAAAAAAAAA_AGGAAAAAA___AUUAUGGCCA_GCGU_UGGGCAUCGUAAA_AAA ..(((((((((...((...............................((((.........))))............................((...))))...))))).))))..... ( -4.06 = -4.38 + 0.32)

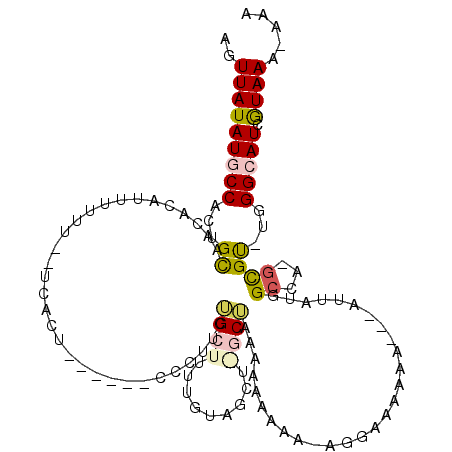
| Location | 17,266,524 – 17,266,635 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.57 |
| Mean single sequence MFE | -20.46 |
| Consensus MFE | -9.67 |
| Energy contribution | -9.63 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17266524 111 - 20766785 UCGCUGGCAAUUGCAUUCAUCCUUCAACAAAUUAAAACAAAGUUAUAUGCCACUGCAACACUUUUUUU--UCACU------CCCUUCAGCUUUUGUAGCUGGCUAAAAAAAAA-AGAGAA ..(((((((...((...........................))....)))))..))....((((((((--(....------....((((((.....))))))......)))))-)))).. ( -17.27) >DroPse_CAF1 23435 109 - 1 UCAGUGGCAAUUGCACUCAUCCUUCAACAAAUUAAAACAAAGUUAUAUGCCACUGUUAAACAUUUUUU-UUUUGUGAGUGCCCCUUCUGCCA----------CAGCAGAAAAAAAGGAAC .((((((((...((...........................))....)))))))).......((((((-((((((..(((.(......).))----------).)))))))))))).... ( -21.63) >DroSec_CAF1 1596 111 - 1 UCGGUGGCAAUUGCAUUCAUCCUUCAACAAAUUAAAACAAAGUUAUAUGCCACUGCAACACUUU-UUU--UCACU------CCCUUCAGCUUUUGUAGCUAGCUAAAAAAAAAGCGAAAA .((((((((...((...........................))....)))))))).........-...--.....------...(((.((((((.(((....)))....))))))))).. ( -18.33) >DroEre_CAF1 1822 111 - 1 UCGGUGGCAAUUGCAUUCAUCCUUCAACAAAUUAAAACAAAGUUAUAUGCCACUGCAACACAUUUUUU--UCACU------CCCUGCUGCUUUUGUAGCUGGCUAAAACAAAA-AGGGAA .((((((((...((...........................))....)))))))).............--....(------(((((((((....)))))((.......))...-))))). ( -23.13) >DroPer_CAF1 23625 110 - 1 UCAGUGGCAAUUGCACUCAUCCUUCAACAAAUUAAAACAAAGUUAUAUGCCACUGUUAAACAUUUUUUUUUUUUUGAGUGCCCCUUCUGCCA----------CAAAAAAAAAAAAAGAAC .((((((((...((...........................))....)))))))).......((((((((((((((.(.((.......))).----------)))))))))))))).... ( -21.93) >consensus UCGGUGGCAAUUGCAUUCAUCCUUCAACAAAUUAAAACAAAGUUAUAUGCCACUGCAACACAUUUUUU__UCACU______CCCUUCUGCUUUUGUAGCU_GCUAAAAAAAAA_AGGAAA .((((((((...((...........................))....))))))))................................................................. ( -9.67 = -9.63 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:19 2006