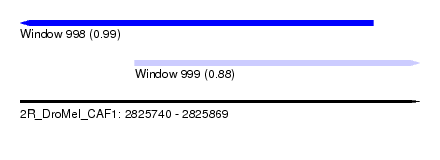
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,825,740 – 2,825,869 |
| Length | 129 |
| Max. P | 0.988782 |

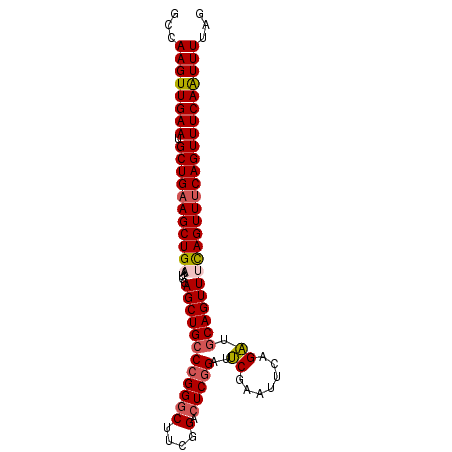
| Location | 2,825,740 – 2,825,854 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 84.12 |
| Mean single sequence MFE | -39.86 |
| Consensus MFE | -28.84 |
| Energy contribution | -29.16 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2825740 114 - 20766785 AAACUGAAACUGCAUCUGAAUUCGAAUCCGAGUCCGAAGCCCGGGCAGCUGCAUUCAGCUACAGCAUUCAACUUGGCAACAGGAAGUGGCAGCGGUAUUGUUGCCGCUUAUCUG ...(((((..((((.((....(((....)))(((((.....))))))).)))))))))...(((.......((((....))))(((((((((((....)))))))))))..))) ( -39.30) >DroSec_CAF1 14767 114 - 1 AAACUGAAACUGCAUCUGAAUCCGAAUCCGAGUCCGAAGCCCGGGCAGCUGCAUACAGCUUCAGCAUUCAACUUGGCAACAGGAAGUGGCAGCGGUAUUGUUGCCACUUAUCUG ....((((..(((..................(((((.....)))))(((((....)))))...))))))).((((....))))(((((((((((....)))))))))))..... ( -38.70) >DroSim_CAF1 27962 114 - 1 AAACUGAAACUGCAUCUGAAUUCGAAUCCGAGUCCGAAGCCCGGGCAGCUGCAUACAGCUUCAGCAUUCAACUUGGCAACAGGAAGUGGCAGCGGUAUUGUUGCCGCUUAUCUG ....((((..(((........(((....)))(((((.....)))))(((((....)))))...))))))).((((....))))(((((((((((....)))))))))))..... ( -39.10) >DroEre_CAF1 14603 89 - 1 AAAAC------------GAAACUGCAGCCGAAUCUUAAGCCCGGGCAGCUGCAUUCAGC-------------UUGGCAACAGGAAGUGGCAGCGUUAUUGUUGCCGCUUAUCUG .....------------.........(((((.......((....)).((((....))))-------------)))))..(((((((((((((((....))))))))))).)))) ( -33.00) >DroYak_CAF1 15087 114 - 1 AAACUAAAACUGAAGCUGAAACUGCAUCUGAUUCCGAAGCCCGGGCAGCUGCAUUCAGCUUCAGCAUUCAACUUGGCAACAGGAAGUGGCAGCGGUAUUGUUGCCGCUUAUCUG .........(((((((((((..((((.(((...(((.....))).))).))))))))))))))).......((((....))))(((((((((((....)))))))))))..... ( -49.20) >consensus AAACUGAAACUGCAUCUGAAUCCGAAUCCGAGUCCGAAGCCCGGGCAGCUGCAUUCAGCUUCAGCAUUCAACUUGGCAACAGGAAGUGGCAGCGGUAUUGUUGCCGCUUAUCUG ...........................(((((((((.....)))))(((((....)))))............))))...(((((((((((((((....))))))))))).)))) (-28.84 = -29.16 + 0.32)

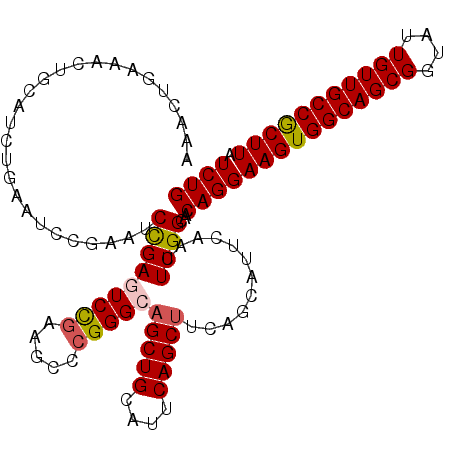
| Location | 2,825,777 – 2,825,869 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 92.57 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -26.00 |
| Energy contribution | -26.87 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2825777 92 + 20766785 GCCAAGUUGAAUGCUGUAGCUGAAUGCAGCUGCCCGGGCUUCGGACUCGGAUUCGAAUUCAGAUGCAGUUUCAGUUUCAGUUUCAAUUUUAG ...((((((((.((((.(((((((....(((((((((((....).)))))..((.......)).)))))))))))).))))))))))))... ( -32.30) >DroSec_CAF1 14804 92 + 1 GCCAAGUUGAAUGCUGAAGCUGUAUGCAGCUGCCCGGGCUUCGGACUCGGAUUCGGAUUCAGAUGCAGUUUCAGUUUCAGUUUCAAUUUUAG ...((((((((.((((((((((..((((.(((.(((((.((((....)))))))))...))).))))....))))))))))))))))))... ( -35.40) >DroSim_CAF1 27999 92 + 1 GCCAAGUUGAAUGCUGAAGCUGUAUGCAGCUGCCCGGGCUUCGGACUCGGAUUCGAAUUCAGAUGCAGUUUCAGUUUCAGUUUCAAUUUUAG ...((((((((.((((((((((..((((.(((.((((((....).))))).........))).))))....))))))))))))))))))... ( -34.60) >DroYak_CAF1 15124 92 + 1 GCCAAGUUGAAUGCUGAAGCUGAAUGCAGCUGCCCGGGCUUCGGAAUCAGAUGCAGUUUCAGCUUCAGUUUUAGUUUCAGUUUCAGUUUGAG ..(((..((((.((((((((((((((((.(((.(((.....)))...))).))))..))))))))))))............))))..))).. ( -32.62) >consensus GCCAAGUUGAAUGCUGAAGCUGAAUGCAGCUGCCCGGGCUUCGGACUCGGAUUCGAAUUCAGAUGCAGUUUCAGUUUCAGUUUCAAUUUUAG ...((((((((.(((((((((((.((((.(((.(((.....)))..(((....)))...))).))))...)))))))))))))))))))... (-26.00 = -26.87 + 0.87)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:05 2006