
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,259,634 – 17,259,737 |
| Length | 103 |
| Max. P | 0.830021 |

| Location | 17,259,634 – 17,259,737 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 72.47 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -8.44 |
| Energy contribution | -8.42 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.29 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17259634 103 + 20766785 ACAUUUAGAUGG--AGUUGCAGGCAAAUGCCAGCGGCCUUCGCAUCUCAGCAC--UGCCAUCCCACUCGA-----GGA-ACUCCUCCAGAUGGA---GCUUUAAUUUAAAACAUCA .......(((((--(((.((.(((....))).(((.....)))......))))--).)))))(((((.((-----((.-...)))).)).))).---................... ( -30.40) >DroSec_CAF1 55850 84 + 1 ACAUUUAGAUGG--AGUUGCAGGC-------------------AUCUCUGCAC--UGCCAUCCCACUCGA-----GGA-ACUCCUCCAGAUGGA---GCUUUAAUUUAAAACAUCA ...(((((((((--((((((((((-------------------(....))).)--)))....(((((.((-----((.-...)))).)).))))---))))).)))))))...... ( -25.50) >DroSim_CAF1 53507 84 + 1 ACAUUUAGAUGG--AGUUGCAGGC-------------------AUCUCUGCAC--UGCCAUCCCACUCGA-----GGA-ACUCCUCCAGAUGGA---GCUUUAAUUUAAAACAUCA ...(((((((((--((((((((((-------------------(....))).)--)))....(((((.((-----((.-...)))).)).))))---))))).)))))))...... ( -25.50) >DroEre_CAF1 52865 100 + 1 ACAUUUAGAUGG--AGUUGCAGGCAAAUGCCAGCGGGCUUCGCAUCUUUUCAC--UGCCAUCCCACUCGA-----GGG-GC---UCCAGAUGGA---GCUUUAAUUUAAAACAUCA .......(((((--(((.((.(((....))).))(((....(((.........--)))...)))))))..-----(((-((---(((....)))---))))).........)))). ( -31.60) >DroYak_CAF1 51968 102 + 1 ACAUUUAGAUGG--AGUUGCAGGCAAAUGCCAGCGGGCUUCGCAUCUCCUCACCAUGCCACGCCACUCGA-----GCG-GC---UCCAGAUGGA---GCUUUAAUUUAAAACAUCA .......(((((--((((((.(((....))).)))(((...((((.........))))...)))))))..-----..(-((---(((....)))---)))...........)))). ( -31.60) >DroAna_CAF1 50568 99 + 1 ACAUUUAGAUGGGAAGUUGCAGGCAAAUGCCAGAGGCCCCAG--------G----CCCCAGCCCACUUGGAGCCAGGGAGU---UGGAGAAAGAGUUGCUUUAAUC--AAGCGUCA .......(((..(((((.((.(((....)))...((((...)--------)----))(((((.(.((((....))))).))---))).......)).))))).)))--........ ( -29.30) >consensus ACAUUUAGAUGG__AGUUGCAGGCAAAUGCCAGCGG_CUUCG_AUCUCUGCAC__UGCCAUCCCACUCGA_____GGA_AC___UCCAGAUGGA___GCUUUAAUUUAAAACAUCA .......((((...(((((.((((....................................(((............)))......(((....)))...))))))))).....)))). ( -8.44 = -8.42 + -0.02)

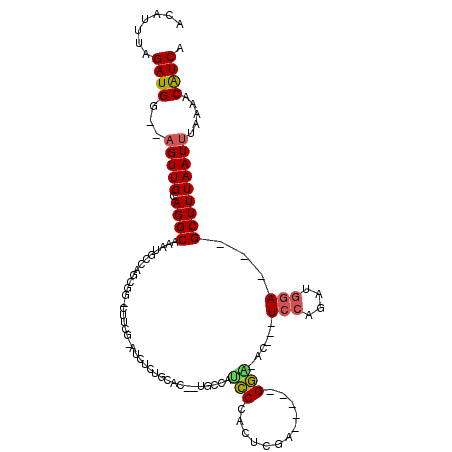
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:11 2006