| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,245,803 – 17,245,901 |
| Length | 98 |
| Max. P | 0.892750 |

| Location | 17,245,803 – 17,245,901 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.76 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -22.30 |
| Energy contribution | -22.63 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
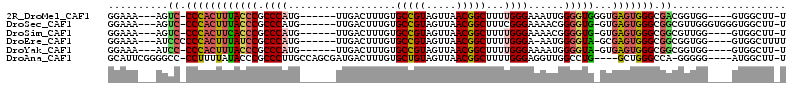
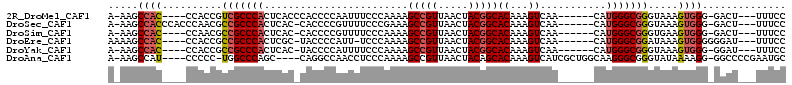
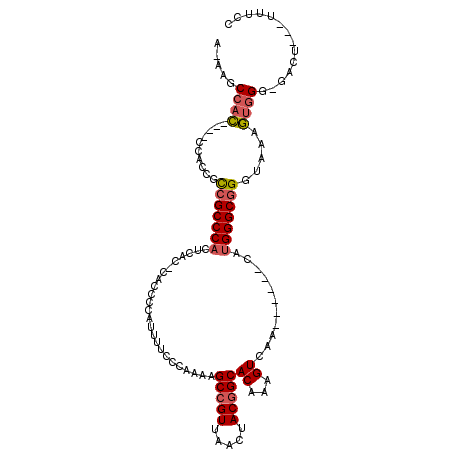
>2R_DroMel_CAF1 17245803 98 + 20766785 GGAAA---AGUC-CCCACUUUACCCGCCCAUG------UUGACUUUGUGCCGUAGUUAACGGCUUUUGGGAAAUUGGGGUGGGUGAGUGGGCGACGGUGG----GUGGCUU-U ...((---((((-((((((((((((((((..(------((..((....(((((.....)))))....))..)))..))))))))))...(....))))))----).)))))-) ( -45.50) >DroSec_CAF1 49717 101 + 1 GGAAA---AGUC-CCCACUUUACCCGCCCAUG------UUGACUUUGUGCCGUAGUUAACGGCUUUCGGGAAAACGGGGUG-GUGAGUGGGCGGCGUUGGGUGGGUGGCUU-U ...((---((((-((((((..((((((((((.------.(.((((((((((((.....)))))(((....)))))))))).-)...)))))))).))..)))))).)))))-) ( -48.70) >DroSim_CAF1 48436 97 + 1 GGAAA---AGUC-CCCACUUCACCCGCCCAUG------UUGACUUUGUGCCGUAGUUAACGGCUUUUGGGAAAACGGGGUG-GUGAGUGGGCGGCGUUGG----GUGGCUU-U ...((---((((-((((....((((((((((.------.(.((((((((((((.....)))))..........))))))).-)...)))))))).)))))----).)))))-) ( -40.40) >DroEre_CAF1 47826 98 + 1 GGAAA---AUCCCCCCACUUUAUCCGCCCAUG------UUGACUUUGUGCCGUAGUUAACGGCUUUUGGGA-AAUGGGGUA-GCGAGUGGGCGGCGGUGG----GUGGCUUUU ..(((---(.((.((((((....((((((((.------...((((..((((((.....)))))........-.)..)))).-....)))))))).)))))----).)).)))) ( -38.60) >DroYak_CAF1 47117 97 + 1 GGAAA---AUCC-CCCACUUUACCCGCCCAUG------UUGACUUUGUGCCGUAGUUAACGGCUUUUGGGAAAAUGGGGUA-GUGAGUGGGCGGCGGUGG----GUGGCUU-U .....---..((-((((((....((((((((.------...((((..((((((.....)))))..........)..)))).-....)))))))).)))))----).))...-. ( -37.50) >DroAna_CAF1 44995 102 + 1 GCAUUCGGGGCC-CCUUUUAUACCCGCCCUUGCCAGCGAUGACUUUGUGCUGUAGUUAACGGCUUUUGGGAGGUUGGCCUG----GCUGGGCCA-GGGGG----AUGGCUU-U .(((((((....-)).......(((((((..(((((.(..((((((.((((((.....)))))....).))))))..))))----)).))))..-)))))----)))....-. ( -38.30) >consensus GGAAA___AGCC_CCCACUUUACCCGCCCAUG______UUGACUUUGUGCCGUAGUUAACGGCUUUUGGGAAAAUGGGGUG_GUGAGUGGGCGGCGGUGG____GUGGCUU_U ..........((.((((((((((((.((((..................(((((.....)))))...))))......)))))...))))))).))................... (-22.30 = -22.63 + 0.34)
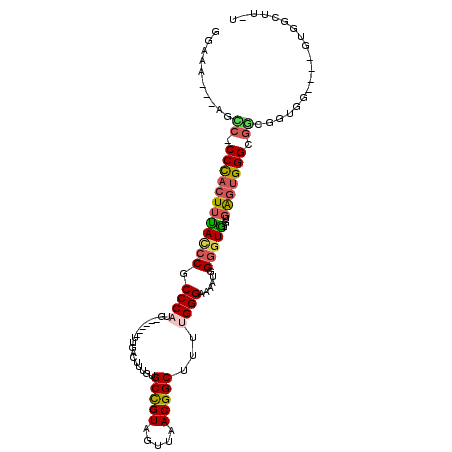
| Location | 17,245,803 – 17,245,901 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.76 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -17.74 |
| Energy contribution | -18.08 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17245803 98 - 20766785 A-AAGCCAC----CCACCGUCGCCCACUCACCCACCCCAAUUUCCCAAAAGCCGUUAACUACGGCACAAAGUCAA------CAUGGGCGGGUAAAGUGGG-GACU---UUUCC .-.......----.....(((.((((((.((((.(((.............(((((.....)))))..........------...))).))))..))))))-))).---..... ( -33.57) >DroSec_CAF1 49717 101 - 1 A-AAGCCACCCACCCAACGCCGCCCACUCAC-CACCCCGUUUUCCCGAAAGCCGUUAACUACGGCACAAAGUCAA------CAUGGGCGGGUAAAGUGGG-GACU---UUUCC (-(((.(.(((((...((.(((((((.....-.....((......))...(((((.....)))))..........------..)))))))))...)))))-).))---))... ( -33.80) >DroSim_CAF1 48436 97 - 1 A-AAGCCAC----CCAACGCCGCCCACUCAC-CACCCCGUUUUCCCAAAAGCCGUUAACUACGGCACAAAGUCAA------CAUGGGCGGGUGAAGUGGG-GACU---UUUCC .-.......----.........(((((((((-(..(((((..........(((((.....)))))..........------.)))))..)))).))))))-....---..... ( -32.70) >DroEre_CAF1 47826 98 - 1 AAAAGCCAC----CCACCGCCGCCCACUCGC-UACCCCAUU-UCCCAAAAGCCGUUAACUACGGCACAAAGUCAA------CAUGGGCGGAUAAAGUGGGGGGAU---UUUCC .....((.(----((((..(((((((.....-.........-........(((((.....))))).....(....------).))))))).....)))))))...---..... ( -32.70) >DroYak_CAF1 47117 97 - 1 A-AAGCCAC----CCACCGCCGCCCACUCAC-UACCCCAUUUUCCCAAAAGCCGUUAACUACGGCACAAAGUCAA------CAUGGGCGGGUAAAGUGGG-GGAU---UUUCC .-...((.(----((((..(((((((.....-..................(((((.....))))).....(....------).))))))).....)))))-))..---..... ( -31.40) >DroAna_CAF1 44995 102 - 1 A-AAGCCAU----CCCCC-UGGCCCAGC----CAGGCCAACCUCCCAAAAGCCGUUAACUACAGCACAAAGUCAUCGCUGGCAAGGGCGGGUAUAAAAGG-GGCCCCGAAUGC .-.......----.((((-(((((....----..)))).(((((((....((.((.....)).)).....((((....))))..))).)))).....)))-)).......... ( -30.60) >consensus A_AAGCCAC____CCACCGCCGCCCACUCAC_CACCCCAUUUUCCCAAAAGCCGUUAACUACGGCACAAAGUCAA______CAUGGGCGGGUAAAGUGGG_GACU___UUUCC .....((((..........(((((((........................(((((.....)))))((...))...........))))))).....)))).............. (-17.74 = -18.08 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:09 2006