| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,234,751 – 17,234,842 |
| Length | 91 |
| Max. P | 0.809648 |

| Location | 17,234,751 – 17,234,842 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 81.34 |
| Mean single sequence MFE | -23.52 |
| Consensus MFE | -12.97 |
| Energy contribution | -13.72 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
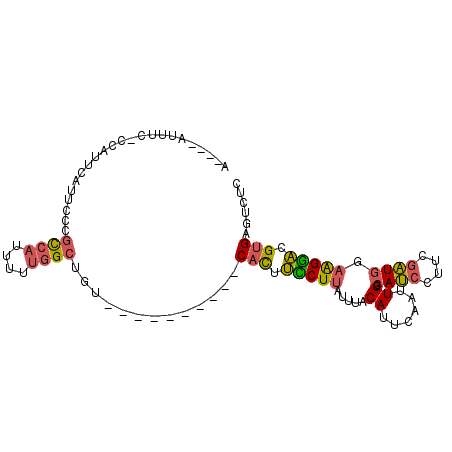
>2R_DroMel_CAF1 17234751 91 + 20766785 GAGACUCACGUCCUUCAAUCGAAGGAUGCCAAUUGAAUGUAAAUAAGAAAGUG----------ACAGCCAAAAAUGGCGGGAAUGAAUGGGGAAAUUCAUU ....(((.((((((((....)))))))).........................----------...((((....)))))))(((((((......))))))) ( -23.10) >DroPse_CAF1 33682 94 + 1 GACUCUCUCUCUCCUCCAAUGAACGCUGCCAAUUGAAUGUAAAUGAGAGAAUGGGUGGCGCUGUCAGCUAAA-CGGAUAGGAAUGGAUUG-A-GAG----U .((((((..(((..(((......(((..((..((..((....))..)).....))..)))((((.......)-)))...)))..)))..)-)-)))----) ( -26.40) >DroSec_CAF1 34421 90 + 1 GAGACUCACGUCCUUCCAUCGAAGGAUGCCAAUUGAAUGUAAAUAAGGAAGUG----------ACAGCCAAAAAUGGCGGGAAUGAAUGG-GAAAUUCAUU ....(((..(((((((((((....)))....(((.......)))..))))).)----------)).((((....)))))))(((((((..-...))))))) ( -24.80) >DroSim_CAF1 37008 90 + 1 GAGACUCACGUCCUUCCAUCGAAGGAUGCCAAUUGAAUGUAAAUAAGGAAGUG----------ACAGCCAAAAAUGGCGGGAAUGAAUGG-GAAAUUCAUU ....(((..(((((((((((....)))....(((.......)))..))))).)----------)).((((....)))))))(((((((..-...))))))) ( -24.80) >DroEre_CAF1 36390 86 + 1 AGGACUCACGUCCUUCCAUCGAAGGAUGCCAAUUGAAUGUAAAUAAGGAAGUG----------ACAGCCAAAAAUGGCGGGAAUGAAUGG-GAAAU----U ....((((.(((((((((((....)))....(((.......)))..))))).)----------)).((((....)))).........)))-)....----. ( -21.00) >DroYak_CAF1 35214 86 + 1 AGGACUCACGUCCUUCCAUCGAAGGAUGCCAAUUGAAUGUAAAUAAGGAAGUG----------ACAGCCAAAAAUGGCGGGAAUGAAUGG-GAAAU----U ....((((.(((((((((((....)))....(((.......)))..))))).)----------)).((((....)))).........)))-)....----. ( -21.00) >consensus GAGACUCACGUCCUUCCAUCGAAGGAUGCCAAUUGAAUGUAAAUAAGGAAGUG__________ACAGCCAAAAAUGGCGGGAAUGAAUGG_GAAAU____U ....(((.((((((((....))))))))......................................((((....))))))).................... (-12.97 = -13.72 + 0.75)

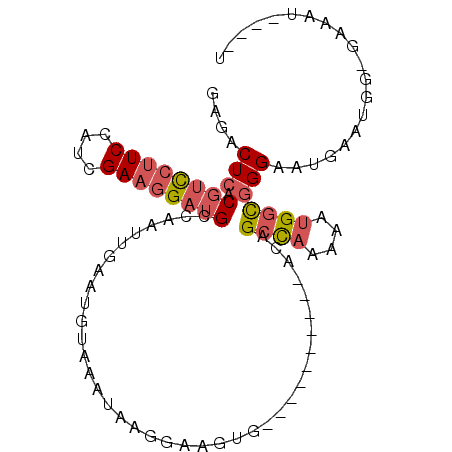
| Location | 17,234,751 – 17,234,842 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 81.34 |
| Mean single sequence MFE | -21.39 |
| Consensus MFE | -11.98 |
| Energy contribution | -11.82 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17234751 91 - 20766785 AAUGAAUUUCCCCAUUCAUUCCCGCCAUUUUUGGCUGU----------CACUUUCUUAUUUACAUUCAAUUGGCAUCCUUCGAUUGAAGGACGUGAGUCUC (((((((......)))))))...((((....))))...----------................(((((((((......)))))))))((((....)))). ( -21.80) >DroPse_CAF1 33682 94 - 1 A----CUC-U-CAAUCCAUUCCUAUCCG-UUUAGCUGACAGCGCCACCCAUUCUCUCAUUUACAUUCAAUUGGCAGCGUUCAUUGGAGGAGAGAGAGAGUC (----(((-(-(..((..((((.....(-((.....)))(((((...(((....................)))..)))))....))))..))..)))))). ( -21.05) >DroSec_CAF1 34421 90 - 1 AAUGAAUUUC-CCAUUCAUUCCCGCCAUUUUUGGCUGU----------CACUUCCUUAUUUACAUUCAAUUGGCAUCCUUCGAUGGAAGGACGUGAGUCUC (((((((...-..)))))))...((((....))))..(----------(((.(((((.....((......)).((((....)))).))))).))))..... ( -21.20) >DroSim_CAF1 37008 90 - 1 AAUGAAUUUC-CCAUUCAUUCCCGCCAUUUUUGGCUGU----------CACUUCCUUAUUUACAUUCAAUUGGCAUCCUUCGAUGGAAGGACGUGAGUCUC (((((((...-..)))))))...((((....))))..(----------(((.(((((.....((......)).((((....)))).))))).))))..... ( -21.20) >DroEre_CAF1 36390 86 - 1 A----AUUUC-CCAUUCAUUCCCGCCAUUUUUGGCUGU----------CACUUCCUUAUUUACAUUCAAUUGGCAUCCUUCGAUGGAAGGACGUGAGUCCU .----.....-(((((.......((((...(((..(((----------.............)))..))).)))).......))))).(((((....))))) ( -21.56) >DroYak_CAF1 35214 86 - 1 A----AUUUC-CCAUUCAUUCCCGCCAUUUUUGGCUGU----------CACUUCCUUAUUUACAUUCAAUUGGCAUCCUUCGAUGGAAGGACGUGAGUCCU .----.....-(((((.......((((...(((..(((----------.............)))..))).)))).......))))).(((((....))))) ( -21.56) >consensus A____AUUUC_CCAUUCAUUCCCGCCAUUUUUGGCUGU__________CACUUCCUUAUUUACAUUCAAUUGGCAUCCUUCGAUGGAAGGACGUGAGUCUC .......................((((....)))).............(((.(((((.....((......)).((((....)))).))))).)))...... (-11.98 = -11.82 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:50 2006