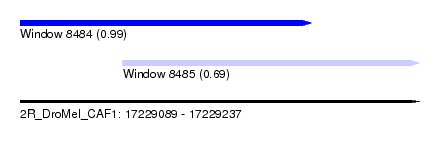
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,229,089 – 17,229,237 |
| Length | 148 |
| Max. P | 0.994530 |

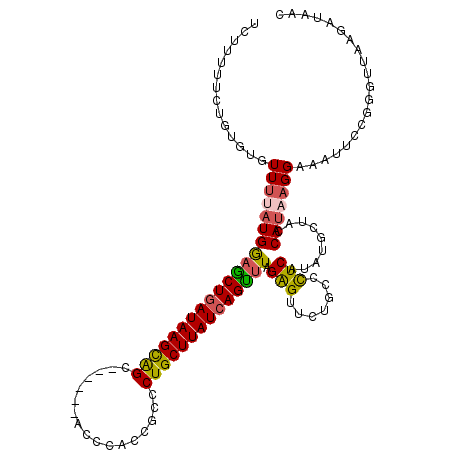
| Location | 17,229,089 – 17,229,197 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.33 |
| Mean single sequence MFE | -24.02 |
| Consensus MFE | -19.83 |
| Energy contribution | -18.99 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17229089 108 + 20766785 AACUGUAACUGUCCUACCUUUAUUACAAAUUUCGAGCUUCUGUUUCUGUGUGUUUUAUGAAGCUGAUAAGCAGC------ACCCACCGCCCUGCUUAUCAGCUUAGAGUUCUGC ...(((((..(........)..)))))......((((((........(((.....))).((((((((((((((.------.(.....)..)))))))))))))).))))))... ( -25.60) >DroSec_CAF1 28894 101 + 1 AACUUUUA-------ACCUUUAUUACAAAUUUCGAACUUCUUUUUCUGUGUGUUUUAUGGAGCUAAUAAGCAGC------ACCCACCGCCCUGCUUAUCAGUUUAGAGUUCUGC ........-------..................((((((.....((((((.....))))))(((.((((((((.------.(.....)..)))))))).)))...))))))... ( -18.40) >DroSim_CAF1 31313 98 + 1 ----------------CCUUUAUUACAACUUUCGAGCUUCUGUUUCUGUGUGUUUUAUGGAACUGAUAAGCGGUUCCAUAACCCACCGCCCUGCUUAUCAGUUUAGAGUUCUGC ----------------.................(((((.(((...(((.(((..(((((((((((.....)))))))))))..))).((...))....)))..))))))))... ( -26.60) >DroEre_CAF1 29472 104 + 1 AGCUUUAAUUUUCCUACCUUUAUUACAAAUGUCGAGGUUUUUCUUUGG---UUUGUUUGGUGCUGAUAAGUAGC------ACCC-CCGCCCUGCUUAUCAGUUUAGAGUUCUGC ((((((((.......(((......((((((..(((((.....))))))---)))))..)))((((((((((((.------.(..-..)..)))))))))))))))))))).... ( -26.60) >DroYak_CAF1 29451 103 + 1 AGCUUUAACCUUCCUACCUUUAUUACAAAUUUCGAGCUUCUUUUUU-----UUUUUAUGAAGCUGAUAAGUAGC------ACCCCCCGCCCUGCUUAUCAGUUUAGGGCUCUGC ((((((((.((.......................((((((......-----.......))))))(((((((((.------.(.....)..))))))))))).)))))))).... ( -22.92) >consensus AACUUUAA___UCCUACCUUUAUUACAAAUUUCGAGCUUCUUUUUCUGUGUGUUUUAUGGAGCUGAUAAGCAGC______ACCCACCGCCCUGCUUAUCAGUUUAGAGUUCUGC .................................((((((....................((((((((((((((.................)))))))))))))).))))))... (-19.83 = -18.99 + -0.84)

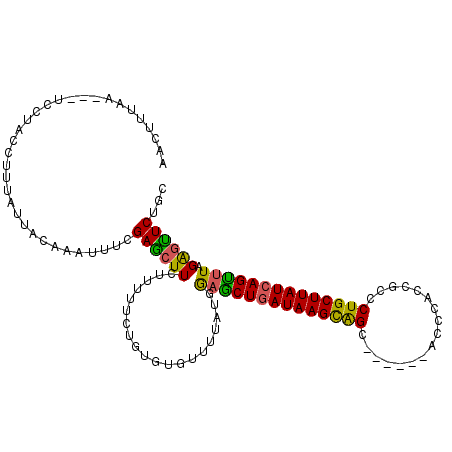
| Location | 17,229,127 – 17,229,237 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.50 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -19.13 |
| Energy contribution | -18.97 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17229127 110 + 20766785 UCUGUUUCUGUGUGUUUUAUGAAGCUGAUAAGCAGC------ACCCACCGCCCUGCUUAUCAGCUUAGAGUUCUGCCCUCAUAUGCUACCAUAAGGAAAUUCCGGGCUAAGAUAAC ............(((((((..((((((((((((((.------.(.....)..))))))))))))))........((((...((((....)))).((.....))))))))))))).. ( -33.50) >DroSec_CAF1 28925 110 + 1 UCUUUUUCUGUGUGUUUUAUGGAGCUAAUAAGCAGC------ACCCACCGCCCUGCUUAUCAGUUUAGAGUUCUGCCUUCAUAUGCUACCAUUAGGAAAUUCCGGGUUAAGAUAAG ((((..((((...........(((((.((((((((.------.(.....)..)))))))).))))).(((((...(((....(((....))).))).)))))))))..)))).... ( -23.10) >DroSim_CAF1 31335 116 + 1 UCUGUUUCUGUGUGUUUUAUGGAACUGAUAAGCGGUUCCAUAACCCACCGCCCUGCUUAUCAGUUUAGAGUUCUGCCUUCAUAUGCUACCAUUAGGAAAUUCCGGGUUAAGAUAAC ......((((.(((..(((((((((((.....)))))))))))..))))((((.((.(((.((..(((....)))..)).))).))........((.....))))))..))).... ( -28.80) >DroEre_CAF1 29510 93 + 1 UUUUCUUUGG---UUUGUUUGGUGCUGAUAAGUAGC------ACCC-CCGCCCUGCUUAUCAGUUUAGAGUUCUGCCCUCAUGUGUUAUCAUAAGGAAA-------------UUAC .(((((((((---(...(((...((((((((((((.------.(..-..)..))))))))))))...)))....))).....(((....))))))))))-------------.... ( -21.60) >DroYak_CAF1 29489 105 + 1 UCUUUUUU-----UUUUUAUGAAGCUGAUAAGUAGC------ACCCCCCGCCCUGCUUAUCAGUUUAGGGCUCUGCCCUCAUGUGUCAGCAUAAGGAAAUUCCGGGUGAAGAUUAC (((((.((-----((((((((((((((((((((((.------.(.....)..))))))))))))))(((((...)))))..........))))))))))........))))).... ( -30.40) >consensus UCUUUUUCUGUGUGUUUUAUGGAGCUGAUAAGCAGC______ACCCACCGCCCUGCUUAUCAGUUUAGAGUUCUGCCCUCAUAUGCUACCAUAAGGAAAUUCCGGGUUAAGAUAAC ..............(((((((((((((((((((((.................)))))))))))))).(((.......))).........))))))).................... (-19.13 = -18.97 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:47 2006