| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
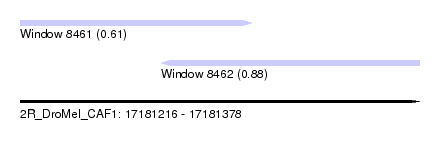
| Location | 17,181,216 – 17,181,378 |
| Length | 162 |
| Max. P | 0.876004 |

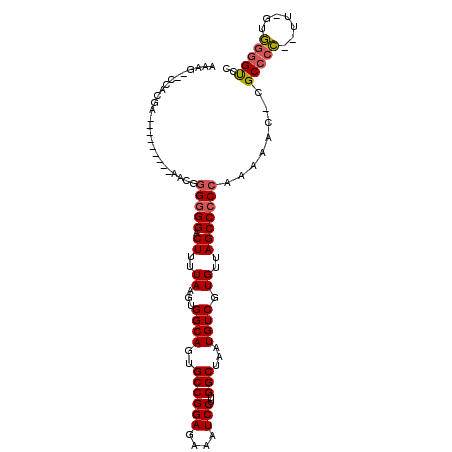
| Location | 17,181,216 – 17,181,310 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.15 |
| Mean single sequence MFE | -36.85 |
| Consensus MFE | -23.56 |
| Energy contribution | -24.95 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612876 |
| Prediction | RNA |
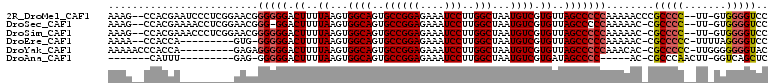
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17181216 94 + 20766785 UGUCUCUAGGAAUUCCCC-GACUUGUGACUCCGUGUGACUCCA------------CUGGGGGUUCGAGUUGGACCCCAC-AA--GGGGCGGGUUUUUGGGGGCUAACACG .((((((((((((((.((-((((((.((((((..(((....))------------)..)))))))))))))).((((..-..--)))).))))))))))))))....... ( -43.30) >DroSec_CAF1 6040 93 + 1 UGUCUCUAGGAAUUCCCC-GACUUGUGACUCCGUGUGGCUCCA------------CUGGGGGUUCGAGUUGGACCCCAC-AA--GGGGCG-GUUUUUGGGGGCUAACACG .(((((((((((((..((-((((((.((((((..(((....))------------)..)))))))))))))).((((..-..--)))).)-))))))))))))....... ( -42.00) >DroSim_CAF1 6034 93 + 1 UGUCUCUAGGAAUUCCCC-GACUUGUGACUCCGUGUGACUCCA------------CUGGGGGUUCGAGUUGGACCCCAC-AA--GGGGCG-GUUUUUGGGGGCUAACACG .(((((((((((((..((-((((((.((((((..(((....))------------)..)))))))))))))).((((..-..--)))).)-))))))))))))....... ( -40.60) >DroEre_CAF1 6087 106 + 1 UGUCUUUAGGAAUUCCCC-GAGUUGUGACUCCGUGUGACUCCGCUUU-UGCUGGACUUGGGGUGCGAGUUGGACCCCUAAAA-GGGGGCG-GUUUUUGGGGGCUAACACG .((((((((((((..(((-((.((((.((((((......(((((...-.)).)))..)))))))))).)))).((((.....-)))))..-))))))))))))....... ( -37.70) >DroYak_CAF1 5902 107 + 1 UGUCUUUAGGAAUUCCCC-GACUUGUGACUCCGUGUGACUCCACUUUCUGCUGGACUUGUGGUGCGAGUUGUACCCCCCCAA-GGGGGCG-GUGUUUGGGGGCUAACACG .....((((....(((((-((...(..((((.(..(.(((((((.....).))))...)).)..)))))..)(((((((...-.)))).)-))..))))))))))).... ( -35.90) >DroAna_CAF1 6923 89 + 1 UGUCUCUUGGAAUUCCCCUGACUUUGGACUCCGU--GACUCCA------------CUUUUUUGUUGAACUGAGCUGACC-AAGUUGGGCG-GU-----GGGGCUAUCACG .(((....((....))...))).........(((--(((((((------------((.....(((.((((.........-.)))).))))-))-----))))...))))) ( -21.60) >consensus UGUCUCUAGGAAUUCCCC_GACUUGUGACUCCGUGUGACUCCA____________CUGGGGGUUCGAGUUGGACCCCAC_AA__GGGGCG_GUUUUUGGGGGCUAACACG .((((((((((((.((((......((.((.....)).))...................(((((((.....))))))).......))))...))))))))))))....... (-23.56 = -24.95 + 1.39)

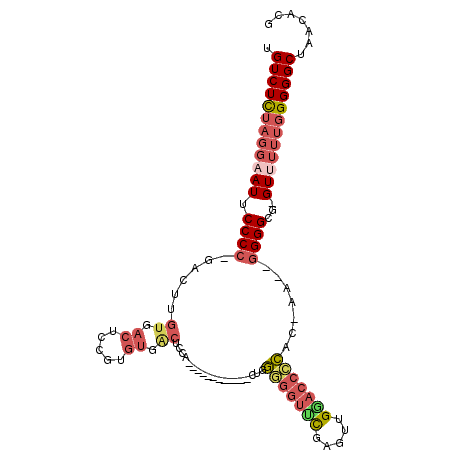
| Location | 17,181,273 – 17,181,378 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.93 |
| Mean single sequence MFE | -36.54 |
| Consensus MFE | -30.03 |
| Energy contribution | -30.28 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17181273 105 - 20766785 AAAG--CCACGAAUCCCUCGGAACGGGGGGACUUUUAAGUGGCAGUGCCGGAGAAAUCCUUGGCUAAUGUCGUGUUAGCCCCCAAAAACCCGCCCC--UU-GUGGGGUCC ...(--((((...((((((......)))))).......)))))......(((....)))..(((((((.....)))))))...........(((((--..-..))))).. ( -39.30) >DroSec_CAF1 6097 103 - 1 AAAG--CCACGAAAACCUCGGAACGGG-GGACUUUUAAGUGGCAGUGCCGGAGAAAUCCUUGGCUAAUGUCGUGUUAGCCCCCAAAAAC-CGCCCC--UU-GUGGGGUCC ....--...(((.....)))....(((-((.((..((...((((..((((((....)))..)))...)))).))..)))))))......-.(((((--..-..))))).. ( -37.00) >DroSim_CAF1 6091 104 - 1 AAAG--CCACGAAACCCUCGGAACGGGGGGACUUUUAAGUGGCAGUGCCGGAGAAAUCCUUGGCUAAUGUCGUGUUAGCCCCCAAAAAC-CGCCCC--UU-GUGGGGUCC ...(--((((....((((((...)))))).........)))))......(((....)))..(((((((.....))))))).........-.(((((--..-..))))).. ( -38.42) >DroEre_CAF1 6155 96 - 1 AAAA--CCACCA---------GUG-GGGGGACUUUUAAGUGGCAGUGCCGGAGAAAUCCUUGGCUAAUGUCGUGUUAGCCCCCAAAAAC-CGCCCCC-UUUUAGGGGUCC ....--......---------...-(((((.((..((...((((..((((((....)))..)))...)))).))..)))))))......-.(((((.-.....))))).. ( -34.40) >DroYak_CAF1 5971 99 - 1 AAAAACCCACCA---------GAGAGGGGGACUUUUAAGUGGCAGUGCCGGAGAAAUCCUUGGCUAAUGUCGUGUUAGCCCCCAAACAC-CGCCCCC-UUGGGGGGGUAC ....((((.((.---------..(((((((..........((((..((((((....)))..)))...))))(((((........)))))-..)))))-))..)))))).. ( -41.70) >DroAna_CAF1 6979 86 - 1 -------CAUUU---------GAG-GGGGGACUUUUAAGUGGCAGUGCCGGAGAAAUCCUUGGCUAAUGUCGUGAUAGCCCC-----AC-CGCCCAACUU-GGUCAGCUC -------...((---------(((-(((((.((.(((...((((..((((((....)))..)))...)))).))).))))))-----.)-)..((.....-))))))... ( -28.40) >consensus AAAG__CCACGA_________AACGGGGGGACUUUUAAGUGGCAGUGCCGGAGAAAUCCUUGGCUAAUGUCGUGUUAGCCCCCAAAAAC_CGCCCC__UU_GUGGGGUCC .........................(((((.((..((...((((..((((((....)))..)))...)))).))..)))))))........(((((.......))))).. (-30.03 = -30.28 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:25 2006