
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,179,290 – 17,179,424 |
| Length | 134 |
| Max. P | 0.952473 |

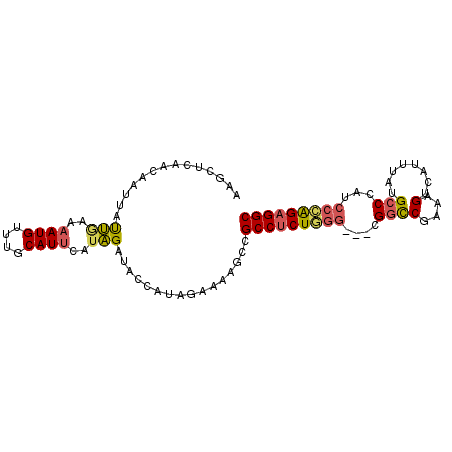
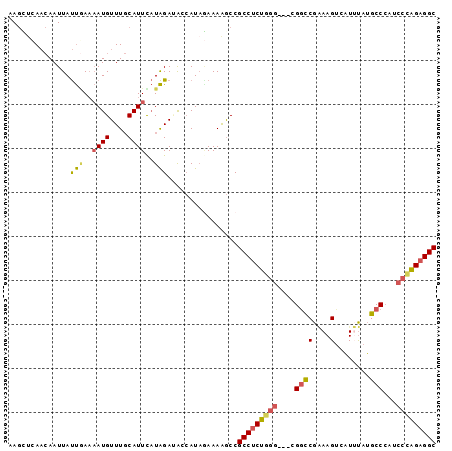
| Location | 17,179,290 – 17,179,384 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 78.15 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -15.82 |
| Energy contribution | -16.46 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17179290 94 + 20766785 AAGUUCAACAAUUAUUGAAAAUGUUUGCAUUCAUAGACACCAUAAAAAAGCCGCCUCUGGG---CGGCCGAAAGUCAUUUAUGCCCAUCCCAGAGGC ...(((((......)))))..((((((......)))))).............(((((((((---.(((..(((....)))..)))...))))))))) ( -25.30) >DroSec_CAF1 4130 94 + 1 AAGCUCAACAAUUAUUCAAAAUGUUUGCAUUCAUAGAUACCAUAGAAAAGCCGCCUCUGGG---CGGCCGAAAGUCAUUUAUGCCCAUCCCAGAGGC ..((..((((.((.....)).)))).))........................(((((((((---.(((..(((....)))..)))...))))))))) ( -23.70) >DroSim_CAF1 4123 94 + 1 AAGCUCAACAAUUAUUGAAAAUGUUUGCAUUCAUAGAUACCAUAGAAAAGCCGCCUCUGGG---CGGCCGAAAGUCAUUUAUGCCCAUCCCAGAGGC ..(((.......((((...((((....))))....)))).........))).(((((((((---.(((..(((....)))..)))...))))))))) ( -24.09) >DroEre_CAF1 4181 94 + 1 AAGGCUAACAAUUAUUGAAAAUGUUUGCAUUUAUAGAGGGCAUACAAAAGCCGCCUCUUGG---CGACCGUAAGUCAUACAUGCCCAUCCCAGAGGC ..((((..((.....))....(((.(((.((....))..))).)))..))))((((((.((---.((..(((.(.....).)))...)))))))))) ( -21.80) >DroAna_CAF1 4936 95 + 1 AAUCCCA--AAUUACCAAUCAUGUUUUCAUUUCUGGAAAAGAUUGAAAGACCGCCCCUUGAAACGGGUCGGAAGAGUUAUCAGCCGAUAAGGGAGGC ..((((.--......(((((...((((((....)))))).))))).....(((.(((.......))).))).....(((((....)))))))))... ( -24.60) >consensus AAGCUCAACAAUUAUUGAAAAUGUUUGCAUUCAUAGAUACCAUAGAAAAGCCGCCUCUGGG___CGGCCGAAAGUCAUUUAUGCCCAUCCCAGAGGC ..............(((..((((....))))..)))................(((((((((....((((....)........)))...))))))))) (-15.82 = -16.46 + 0.64)

| Location | 17,179,290 – 17,179,384 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 78.15 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -16.77 |
| Energy contribution | -17.89 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17179290 94 - 20766785 GCCUCUGGGAUGGGCAUAAAUGACUUUCGGCCG---CCCAGAGGCGGCUUUUUUAUGGUGUCUAUGAAUGCAAACAUUUUCAAUAAUUGUUGAACUU (((((((((.(((.(.............).)))---))))))))).((...(((((((...))))))).)).......((((((....))))))... ( -31.52) >DroSec_CAF1 4130 94 - 1 GCCUCUGGGAUGGGCAUAAAUGACUUUCGGCCG---CCCAGAGGCGGCUUUUCUAUGGUAUCUAUGAAUGCAAACAUUUUGAAUAAUUGUUGAGCUU (((((((((.(((.(.............).)))---)))))))))(((((..(....((.((...(((((....))))).))))....)..))))). ( -31.02) >DroSim_CAF1 4123 94 - 1 GCCUCUGGGAUGGGCAUAAAUGACUUUCGGCCG---CCCAGAGGCGGCUUUUCUAUGGUAUCUAUGAAUGCAAACAUUUUCAAUAAUUGUUGAGCUU (((((((((.(((.(.............).)))---)))))))))(((((....(..((......(((((....)))))......))..).))))). ( -30.82) >DroEre_CAF1 4181 94 - 1 GCCUCUGGGAUGGGCAUGUAUGACUUACGGUCG---CCAAGAGGCGGCUUUUGUAUGCCCUCUAUAAAUGCAAACAUUUUCAAUAAUUGUUAGCCUU .......(((.(((((((((.....)))(((((---((....))))))).....)))))))))......((.((((.((.....)).)))).))... ( -27.30) >DroAna_CAF1 4936 95 - 1 GCCUCCCUUAUCGGCUGAUAACUCUUCCGACCCGUUUCAAGGGGCGGUCUUUCAAUCUUUUCCAGAAAUGAAAACAUGAUUGGUAAUU--UGGGAUU ...((((...((((..((....))..)))).((((((....))))))....((((((.(((.((....)).)))...)))))).....--.)))).. ( -22.70) >consensus GCCUCUGGGAUGGGCAUAAAUGACUUUCGGCCG___CCCAGAGGCGGCUUUUCUAUGGUAUCUAUGAAUGCAAACAUUUUCAAUAAUUGUUGAGCUU (((((((((...(((..............)))....)))))))))(((((...............(((((....)))))............))))). (-16.77 = -17.89 + 1.12)

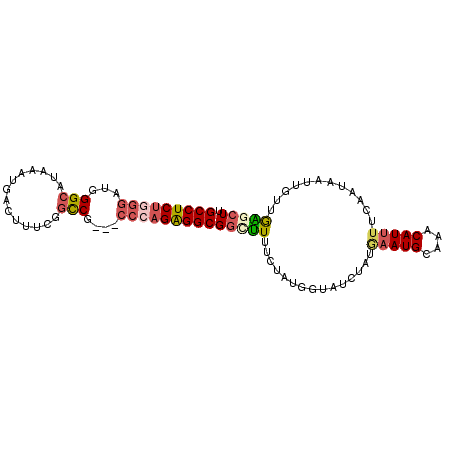
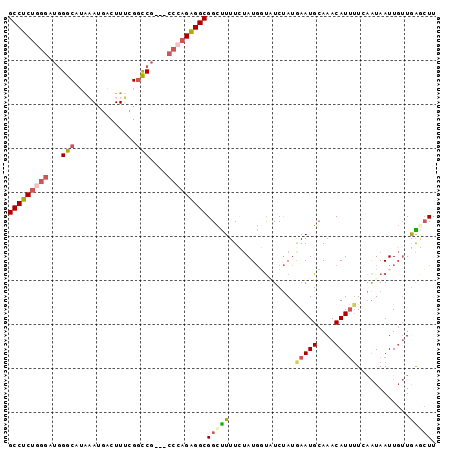
| Location | 17,179,307 – 17,179,424 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.09 |
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -24.44 |
| Energy contribution | -26.68 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17179307 117 + 20766785 AAAAUGUUUGCAUUCAUAGACACCAUAAAAAAGCCGCCUCUGGG---CGGCCGAAAGUCAUUUAUGCCCAUCCCAGAGGCCAAGUUAGUUGACGAUGUCGUCAGCUACAUCGCCAAGCCA .....((((((........................(((((((((---.(((..(((....)))..)))...)))))))))...((.((((((((....))))))))))...).))))).. ( -38.70) >DroSec_CAF1 4147 117 + 1 AAAAUGUUUGCAUUCAUAGAUACCAUAGAAAAGCCGCCUCUGGG---CGGCCGAAAGUCAUUUAUGCCCAUCCCAGAGGCCAAGUUAGUUGACGAUGUCGUCAGCUACAUCGCCAAACCA .....((((((........................(((((((((---.(((..(((....)))..)))...)))))))))...((.((((((((....))))))))))...).))))).. ( -38.70) >DroSim_CAF1 4140 117 + 1 AAAAUGUUUGCAUUCAUAGAUACCAUAGAAAAGCCGCCUCUGGG---CGGCCGAAAGUCAUUUAUGCCCAUCCCAGAGGCCAAGUUAGUUGACGACGUCGUCAGCUACAUCGCCAAACCG .....((((((........................(((((((((---.(((..(((....)))..)))...)))))))))...((.((((((((....))))))))))...).))))).. ( -38.70) >DroEre_CAF1 4198 117 + 1 AAAAUGUUUGCAUUUAUAGAGGGCAUACAAAAGCCGCCUCUUGG---CGACCGUAAGUCAUACAUGCCCAUCCCAGAGGCCCAGUUAGUCGACGAUGUCGCGAGCUACAUCGCCAAACCA .(((((....))))).....((((((.........(((....))---)(((.....)))....))))))........(((...(((....)))((((((....)..))))))))...... ( -31.00) >DroAna_CAF1 4951 120 + 1 AUCAUGUUUUCAUUUCUGGAAAAGAUUGAAAGACCGCCCCUUGAAACGGGUCGGAAGAGUUAUCAGCCGAUAAGGGAGGCCGAGUACAUUAGCGACAUCGAGAGUCAUCUGGCCCUUCCG .((...((((((..(((.....))).)))))).(((.(((.......))).)))..)).(((((....)))))((((((((.((...(((..((....))..)))...)))).)))))). ( -32.30) >consensus AAAAUGUUUGCAUUCAUAGAUACCAUAGAAAAGCCGCCUCUGGG___CGGCCGAAAGUCAUUUAUGCCCAUCCCAGAGGCCAAGUUAGUUGACGAUGUCGUCAGCUACAUCGCCAAACCA .....((((((........................(((((((((....((((....)........)))...)))))))))...((.((((((((....))))))))))...).))))).. (-24.44 = -26.68 + 2.24)

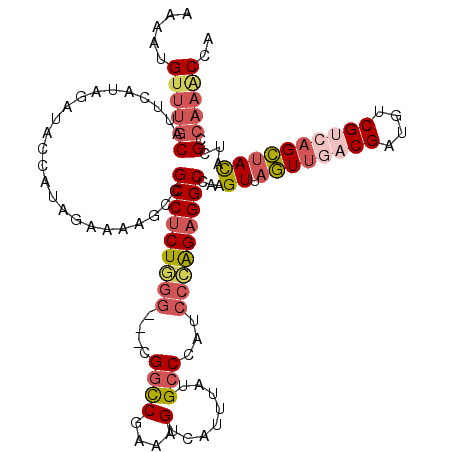
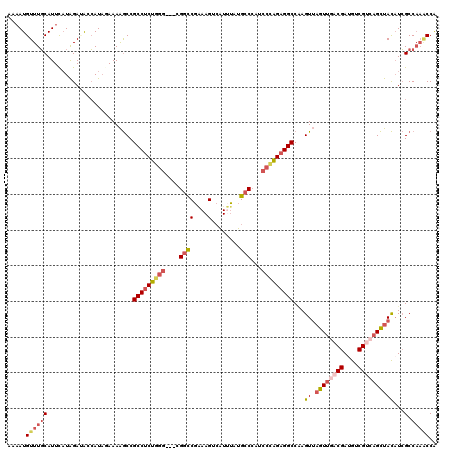
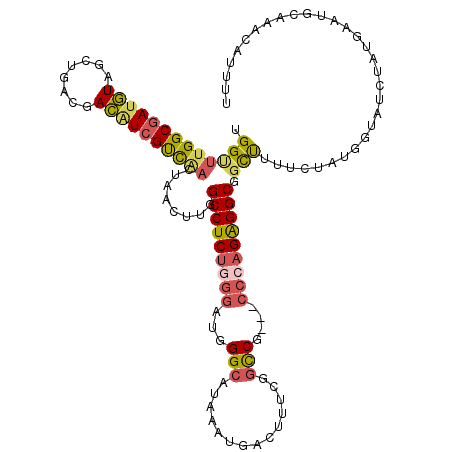
| Location | 17,179,307 – 17,179,424 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.09 |
| Mean single sequence MFE | -39.13 |
| Consensus MFE | -26.66 |
| Energy contribution | -27.22 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17179307 117 - 20766785 UGGCUUGGCGAUGUAGCUGACGACAUCGUCAACUAACUUGGCCUCUGGGAUGGGCAUAAAUGACUUUCGGCCG---CCCAGAGGCGGCUUUUUUAUGGUGUCUAUGAAUGCAAACAUUUU (((.((((((((((........))))))))))))).....(((((((((.(((.(.............).)))---))))))))).((...(((((((...))))))).))......... ( -42.72) >DroSec_CAF1 4147 117 - 1 UGGUUUGGCGAUGUAGCUGACGACAUCGUCAACUAACUUGGCCUCUGGGAUGGGCAUAAAUGACUUUCGGCCG---CCCAGAGGCGGCUUUUCUAUGGUAUCUAUGAAUGCAAACAUUUU (((((.((((((((........))))))))))))).....(((((((((.(((.(.............).)))---))))))))).((..(((.((((...))))))).))......... ( -41.12) >DroSim_CAF1 4140 117 - 1 CGGUUUGGCGAUGUAGCUGACGACGUCGUCAACUAACUUGGCCUCUGGGAUGGGCAUAAAUGACUUUCGGCCG---CCCAGAGGCGGCUUUUCUAUGGUAUCUAUGAAUGCAAACAUUUU .((((.((((((((........))))))))))))......(((((((((.(((.(.............).)))---))))))))).((..(((.((((...))))))).))......... ( -39.92) >DroEre_CAF1 4198 117 - 1 UGGUUUGGCGAUGUAGCUCGCGACAUCGUCGACUAACUGGGCCUCUGGGAUGGGCAUGUAUGACUUACGGUCG---CCAAGAGGCGGCUUUUGUAUGCCCUCUAUAAAUGCAAACAUUUU (((((.((((((((.(....).)))))))))))))..((.((.....(((.(((((((((.....)))(((((---((....))))))).....)))))))))......))...)).... ( -40.40) >DroAna_CAF1 4951 120 - 1 CGGAAGGGCCAGAUGACUCUCGAUGUCGCUAAUGUACUCGGCCUCCCUUAUCGGCUGAUAACUCUUCCGACCCGUUUCAAGGGGCGGUCUUUCAAUCUUUUCCAGAAAUGAAAACAUGAU ((((((((..((.((((.......)))))).......((((((.........))))))...))))))))..((((((....))))))....(((...(((((.(....))))))..))). ( -31.50) >consensus UGGUUUGGCGAUGUAGCUGACGACAUCGUCAACUAACUUGGCCUCUGGGAUGGGCAUAAAUGACUUUCGGCCG___CCCAGAGGCGGCUUUUCUAUGGUAUCUAUGAAUGCAAACAUUUU .(((((((((((((........))))))))))........(((((((((...(((..............)))....))))))))).)))............................... (-26.66 = -27.22 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:21 2006