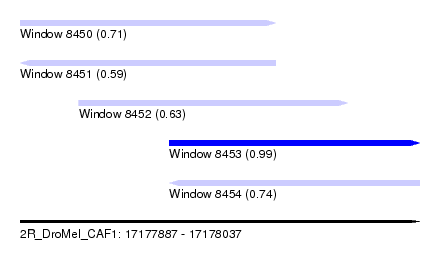
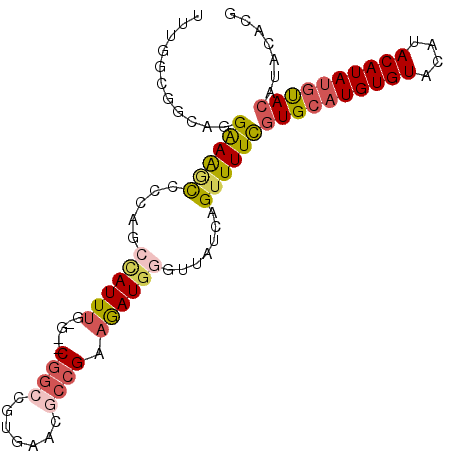
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,177,887 – 17,178,037 |
| Length | 150 |
| Max. P | 0.988983 |

| Location | 17,177,887 – 17,177,983 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 81.10 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -17.84 |
| Energy contribution | -18.73 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
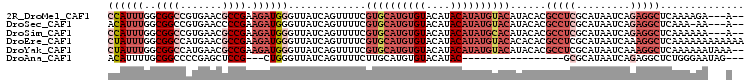
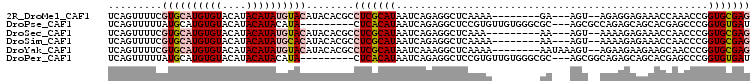
>2R_DroMel_CAF1 17177887 96 + 20766785 AUUGGCGACAGGAAAGCUCCAGCCAUUUG-G--CGGCCGUGAACGCCGAAGAUGGGUUAUCAGUUUUCGUGCAUGUGUACAUACAUAUGUACAUACACG .(((....)))(((((((..((((.((((-(--((........)))))))....))))...)))))))((((((((((....))))))))))....... ( -35.60) >DroSec_CAF1 2748 96 + 1 UUUGGCGGCAGGAAGGCCCCAGACAUUUG-G--CGGCCGUGAACCCCGAAGAUGGGUUAUCAGUUUUCGUGCAUGUGUACAUACAUAUGUACAUACACG .......(((.(((((((((((....)))-)--.))))...(((((.......))))).......))).))).(((((((((....))))))))).... ( -33.30) >DroSim_CAF1 2728 96 + 1 UUUGGCGGCAGGAAAGCCCCAACCAUUUG-G--CGGCCGUGAACGCCGAAGAUGGGUUAUCAGUUUUCGUGCAUGUGUACAUACAUAUGCACAUACACG .((((.(((......)))))))((((((.-.--((((.(....))))).)))))).............((((((((((....))))))))))....... ( -37.80) >DroEre_CAF1 2732 96 + 1 UUUGGCGGCAGGAAACCCACCGCUAUUUG-G--CGGCCAUGAACGCCGAAGAUGGGUUAUCAGUUUUCGUGCAUGUGUACAUACAUAUGUACACACACG ..((((((..(....)...))))))...(-(--((........))))((((((((....))).)))))(((..(((((((((....)))))))))))). ( -34.20) >DroYak_CAF1 2727 96 + 1 GUUGGCGGCAGGAAAACCACCGCUAUUUG-G--CGGCCAUGAACGCCGAAGAUGGGUUAUCAGUUUUCGUGCAUGUGUACAUACAUAUGUACAUACACG ..((((((..((....)).))))))...(-(--((........))))((((((((....))).)))))((((((((((....))))))))))....... ( -32.70) >DroPer_CAF1 9632 80 + 1 ---AGC-----GGAAGUGGUCAAUAUUUCGGCUCUGGCGUGAAC--CGACAAU-GGUUAUCAGUUUUUAUGCAUGUGUACAUACAUACAUA-------- ---(((-----.((((((.....)))))).)))((((....(((--(......-)))).))))....((((.(((((....))))).))))-------- ( -17.20) >consensus UUUGGCGGCAGGAAAGCCCCAGCCAUUUG_G__CGGCCGUGAACGCCGAAGAUGGGUUAUCAGUUUUCGUGCAUGUGUACAUACAUAUGUACAUACACG ...........((((((.....((((((.....((((.......)))).)))))).......))))))((((((((((....))))))))))....... (-17.84 = -18.73 + 0.89)

| Location | 17,177,887 – 17,177,983 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 81.10 |
| Mean single sequence MFE | -30.43 |
| Consensus MFE | -13.35 |
| Energy contribution | -15.10 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17177887 96 - 20766785 CGUGUAUGUACAUAUGUAUGUACACAUGCACGAAAACUGAUAACCCAUCUUCGGCGUUCACGGCCG--C-CAAAUGGCUGGAGCUUUCCUGUCGCCAAU .((((((((((....))))))))))..((.((((...((......))..))))))((((.((((((--.-....))))))))))............... ( -29.40) >DroSec_CAF1 2748 96 - 1 CGUGUAUGUACAUAUGUAUGUACACAUGCACGAAAACUGAUAACCCAUCUUCGGGGUUCACGGCCG--C-CAAAUGUCUGGGGCCUUCCUGCCGCCAAA .((((((((((....))))))))))..(((.(((...(((...(((......)))..))).((((.--(-((......)))))))))).)))....... ( -33.90) >DroSim_CAF1 2728 96 - 1 CGUGUAUGUGCAUAUGUAUGUACACAUGCACGAAAACUGAUAACCCAUCUUCGGCGUUCACGGCCG--C-CAAAUGGUUGGGGCUUUCCUGCCGCCAAA ((((((((((.(((....))).))))))))))............((((...((((.......))))--.-...))))(((((((......))).)))). ( -33.20) >DroEre_CAF1 2732 96 - 1 CGUGUGUGUACAUAUGUAUGUACACAUGCACGAAAACUGAUAACCCAUCUUCGGCGUUCAUGGCCG--C-CAAAUAGCGGUGGGUUUCCUGCCGCCAAA .((((((((((((....))))))))))))..(((..((((..........))))..))).((((.(--(-......))((..((...))..)))))).. ( -35.20) >DroYak_CAF1 2727 96 - 1 CGUGUAUGUACAUAUGUAUGUACACAUGCACGAAAACUGAUAACCCAUCUUCGGCGUUCAUGGCCG--C-CAAAUAGCGGUGGUUUUCCUGCCGCCAAC .((((((((((....))))))))))..(((.(((((((...(((((......)).)))....((((--(-......)))))))))))).)))....... ( -32.00) >DroPer_CAF1 9632 80 - 1 --------UAUGUAUGUAUGUACACAUGCAUAAAAACUGAUAACC-AUUGUCG--GUUCACGCCAGAGCCGAAAUAUUGACCACUUCC-----GCU--- --------(((((((((......)))))))))............(-(...(((--((((......))))))).....)).........-----...--- ( -18.90) >consensus CGUGUAUGUACAUAUGUAUGUACACAUGCACGAAAACUGAUAACCCAUCUUCGGCGUUCACGGCCG__C_CAAAUAGCGGGGGCUUUCCUGCCGCCAAA .((((((((((....))))))))))..((..(((..((((..........))))..)))......................(((......))))).... (-13.35 = -15.10 + 1.75)
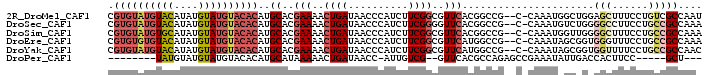
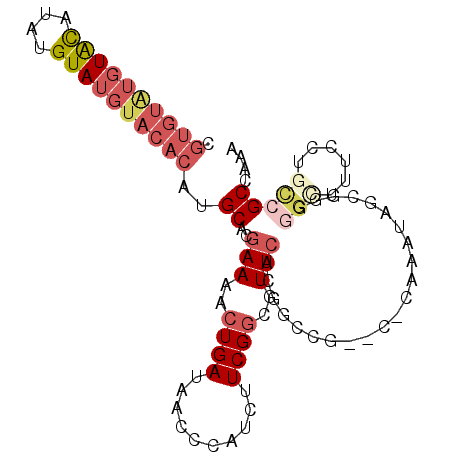
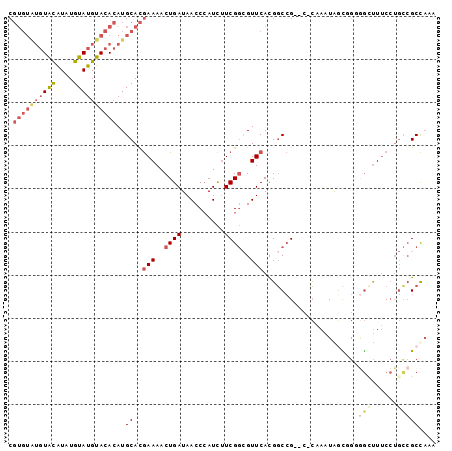
| Location | 17,177,909 – 17,178,010 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 83.54 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -19.26 |
| Energy contribution | -22.43 |
| Covariance contribution | 3.17 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17177909 101 + 20766785 CCAUUUGGCGGCCGUGAACGCCGAAGAUGGGUUAUCAGUUUUCGUGCAUGUGUACAUACAUAUGUACAUACACGCCUCGCAUAAUCAGAGGCUCAAAAGA---A-- ((((((..((((.(....))))).))))))....((.......((((((((((....))))))))))......(((((.........)))))......))---.-- ( -35.30) >DroSec_CAF1 2770 100 + 1 ACAUUUGGCGGCCGUGAACCCCGAAGAUGGGUUAUCAGUUUUCGUGCAUGUGUACAUACAUAUGUACAUACACGCCUCGCAUAAUCAGAGGCUCAAA-AA---A-- ...(((((.....((((((((.......)))))..........((((((((((....))))))))))...)))(((((.........))))))))))-..---.-- ( -31.00) >DroSim_CAF1 2750 101 + 1 CCAUUUGGCGGCCGUGAACGCCGAAGAUGGGUUAUCAGUUUUCGUGCAUGUGUACAUACAUAUGCACAUACACGCCUCGCAUAAUCAGAGGCUCAAAAAA---A-- ((((((..((((.(....))))).)))))).............((((((((((....))))))))))......(((((.........)))))........---.-- ( -37.00) >DroEre_CAF1 2754 106 + 1 CUAUUUGGCGGCCAUGAACGCCGAAGAUGGGUUAUCAGUUUUCGUGCAUGUGUACAUACAUAUGUACACACACGCCUCGCAUAAUCAAAGGCUCAAAAAAAAAAAA ...(((((((........)))))))..((((((..........(((..(((((((((....))))))))))))((...)).........))))))........... ( -28.20) >DroYak_CAF1 2749 104 + 1 CUAUUUGGCGGCCAUGAACGCCGAAGAUGGGUUAUCAGUUUUCGUGCAUGUGUACAUACAUAUGUACAUACACGCCUCGCAUAAUCAAAGGCUCAAAAAAUAAA-- ...(((((((........)))))))..((((((..........((((((((((....))))))))))......((...)).........)))))).........-- ( -27.50) >DroAna_CAF1 3522 83 + 1 ACAUUUUGCGGCCCCGAGCUCCG---CUGGGUUAUCAGUUUUCUUGCAUGUGUACAUAC-----------------GCGCAUAAUCAGAGGCUCUGGGAAUAG--- ...........(((.((((..((---((((....)))))........((((((......-----------------)))))).....)..)))).))).....--- ( -22.70) >consensus CCAUUUGGCGGCCGUGAACGCCGAAGAUGGGUUAUCAGUUUUCGUGCAUGUGUACAUACAUAUGUACAUACACGCCUCGCAUAAUCAGAGGCUCAAAAAA___A__ ((((((..((((.......)))).)))))).............((((((((((....))))))))))......(((((.........))))).............. (-19.26 = -22.43 + 3.17)
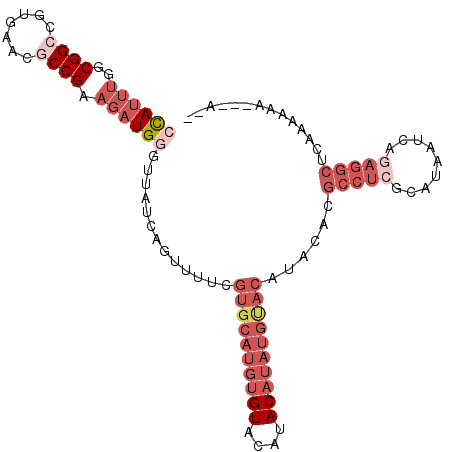

| Location | 17,177,943 – 17,178,037 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 74.57 |
| Mean single sequence MFE | -28.56 |
| Consensus MFE | -13.57 |
| Energy contribution | -14.32 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
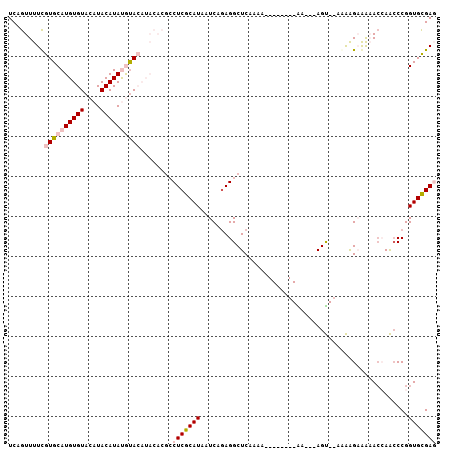
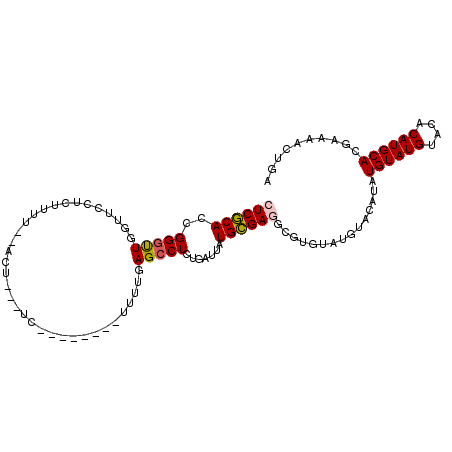
>2R_DroMel_CAF1 17177943 94 + 20766785 UCAGUUUUCGUGCAUGUGUACAUACAUAUGUACAUACACGCCUCGCAUAAUCAGAGGCUCAAAA--------GA---AGU--AGAGGAGAAACCAAACCGGUGCGAG ((..(((((((((((((((....)))))))))).(((..(((((.........)))))((....--------))---.))--)...)))))(((.....)))..)). ( -25.90) >DroPse_CAF1 8535 95 + 1 UCAGUUUUUAUGCAUGUGUACAUACAUACAUA---------CUCACAUAAUCAGAGGCUCCGUGUUGUGGGCGC---AGCGCCAGAGCAGCACGAGCCCGGUGUGAU ........((((.(((((....))))).))))---------.((((((.....(.(((((.(((((((.((((.---..))))...))))))))))))).)))))). ( -36.10) >DroSec_CAF1 2804 93 + 1 UCAGUUUUCGUGCAUGUGUACAUACAUAUGUACAUACACGCCUCGCAUAAUCAGAGGCUCAAA---------AA---AGU--AAAAGAGAAACCAACCCGGUGCGAG ((..(((((((((((((((....))))))))))......(((((.........))))).....---------..---...--....)))))(((.....)))..)). ( -26.00) >DroSim_CAF1 2784 94 + 1 UCAGUUUUCGUGCAUGUGUACAUACAUAUGCACAUACACGCCUCGCAUAAUCAGAGGCUCAAAA--------AA---AGU--AAAAGAGAAACCAACCCGGUGCGAG ((..(((((((((((((((....))))))))))......(((((.........)))))......--------..---...--....)))))(((.....)))..)). ( -28.00) >DroYak_CAF1 2783 97 + 1 UCAGUUUUCGUGCAUGUGUACAUACAUAUGUACAUACACGCCUCGCAUAAUCAAAGGCUCAAAA--------AAUAAAGU--AGAAGAAGAAGCAACCCGGUGCGAG .........((((((((((....))))))))))........(((((((........((((....--------........--.......).)))......))))))) ( -22.65) >DroPer_CAF1 9680 95 + 1 UCAGUUUUUAUGCAUGUGUACAUACAUACAUA---------CUCACAUAAUCAGAGGCUCCGUGUUGUGGGCGC---AGCGGCAGAGCAGCACGAGCCCGGUGUGAU ........((((.(((((....))))).))))---------.((((((.....(.(((((.(((((((..((..---....))...))))))))))))).)))))). ( -32.70) >consensus UCAGUUUUCGUGCAUGUGUACAUACAUAUGUACAUACACGCCUCGCAUAAUCAGAGGCUCAAAA________AA___AGU__AAAAGAAAAACCAACCCGGUGCGAG .........((((((((((....))))))))))........(((((((....................................................))))))) (-13.57 = -14.32 + 0.75)

| Location | 17,177,943 – 17,178,037 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 74.57 |
| Mean single sequence MFE | -27.56 |
| Consensus MFE | -12.55 |
| Energy contribution | -12.38 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
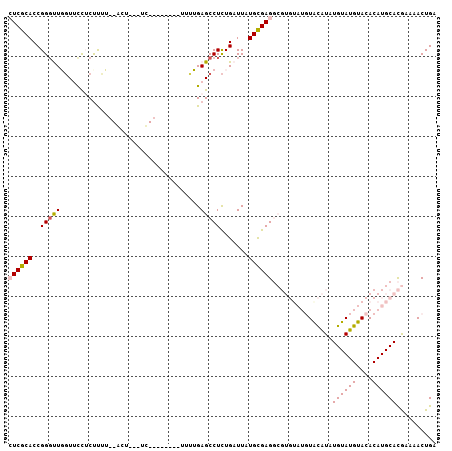
>2R_DroMel_CAF1 17177943 94 - 20766785 CUCGCACCGGUUUGGUUUCUCCUCU--ACU---UC--------UUUUGAGCCUCUGAUUAUGCGAGGCGUGUAUGUACAUAUGUAUGUACACAUGCACGAAAACUGA ((((((.(((...(((((.......--...---..--------....))))).)))....)))))).(((((((((((((....)))))))..))))))........ ( -26.19) >DroPse_CAF1 8535 95 - 1 AUCACACCGGGCUCGUGCUGCUCUGGCGCU---GCGCCCACAACACGGAGCCUCUGAUUAUGUGAG---------UAUGUAUGUAUGUACACAUGCAUAAAAACUGA .(((((..((((.((((((.....))))).---).))))......(((.....)))....))))).---------(((((((((......)))))))))........ ( -30.60) >DroSec_CAF1 2804 93 - 1 CUCGCACCGGGUUGGUUUCUCUUUU--ACU---UU---------UUUGAGCCUCUGAUUAUGCGAGGCGUGUAUGUACAUAUGUAUGUACACAUGCACGAAAACUGA ((((((...(((..(...(((....--...---..---------...)))...)..))).)))))).(((((((((((((....)))))))..))))))........ ( -28.20) >DroSim_CAF1 2784 94 - 1 CUCGCACCGGGUUGGUUUCUCUUUU--ACU---UU--------UUUUGAGCCUCUGAUUAUGCGAGGCGUGUAUGUGCAUAUGUAUGUACACAUGCACGAAAACUGA ((((((...(((..(...(((....--...---..--------....)))...)..))).)))))).((((((((((.(((....))).))))))))))........ ( -29.52) >DroYak_CAF1 2783 97 - 1 CUCGCACCGGGUUGCUUCUUCUUCU--ACUUUAUU--------UUUUGAGCCUUUGAUUAUGCGAGGCGUGUAUGUACAUAUGUAUGUACACAUGCACGAAAACUGA ((((((...((((((((........--........--------....))))....)))).)))))).(((((((((((((....)))))))..))))))........ ( -22.95) >DroPer_CAF1 9680 95 - 1 AUCACACCGGGCUCGUGCUGCUCUGCCGCU---GCGCCCACAACACGGAGCCUCUGAUUAUGUGAG---------UAUGUAUGUAUGUACACAUGCAUAAAAACUGA .(((((..((((.((.((.((...)).)))---).))))......(((.....)))....))))).---------(((((((((......)))))))))........ ( -27.90) >consensus CUCGCACCGGGUUGGUUCCUCUUUU__ACU___UC________UUUUGAGCCUCUGAUUAUGCGAGGCGUGUAUGUACAUAUGUAUGUACACAUGCACGAAAACUGA ((((((..(((((...................................))))).......))))))...............((((((....)))))).......... (-12.55 = -12.38 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:18 2006