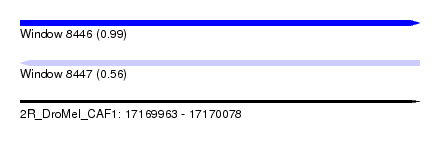
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,169,963 – 17,170,078 |
| Length | 115 |
| Max. P | 0.991375 |

| Location | 17,169,963 – 17,170,078 |
|---|---|
| Length | 115 |
| Sequences | 6 |
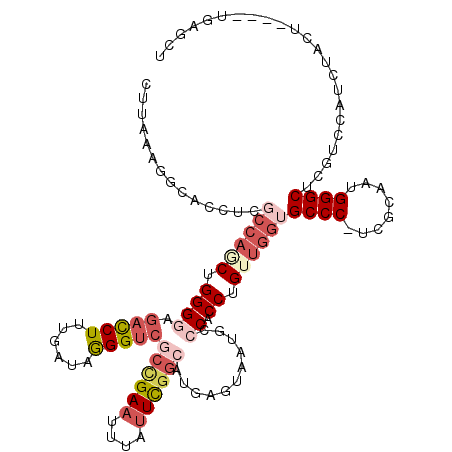
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -39.63 |
| Consensus MFE | -23.27 |
| Energy contribution | -25.22 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17169963 115 + 20766785 CUUAAAGGCAUCUCGCCAUCUGGGGAGACCUUUGAUUGGGUCGCCGAAUUUAUUCGGCAUGAGUAAUGUCCACCUGUUGGUGCCC-UCGCAAUGGGCUCGUCCAUCGACU----UGAGCU .(((((((..((((.((....)).)))))))))))(..((((((((((....))))))((((((......((((....)))).((-.......)))))))).....))))----..)... ( -42.60) >DroSec_CAF1 37124 117 + 1 CUUAAAGGCACCUUGCCAGCUGGGGAGACCUUUGAUUGGGUCGCCGAAUUUAUUCGGCAUGAGUAAUAACCACCUGUUGGUGCCC-UCAUAAUGGGCUCGUCCAUCUACUAC--UGAGCU ......(((.....((((((.((((.(((((......)))))((((((....))))))...........)).)).))))))((((-.......))))...............--...))) ( -40.40) >DroSim_CAF1 38191 115 + 1 CUUAAAGGCACCUUGCCAGCUGGGGAGACCUUUGAUUGGGUCGCCGAAUUUAUUCGGCAUGAGUAAUAACCACCUGUUGGUGCCC-UCAUAAUGGGCUCGUCCAUCUGCU----UGAGCU .....(((((....((((((.((((.(((((......)))))((((((....))))))...........)).)).))))))((((-.......)))).........))))----)..... ( -42.00) >DroEre_CAF1 38107 111 + 1 UUUUAAGCCACUUCGCCAACUGGGCAGACCUUUGAUAGGGUCGCCGAAUUUAUUUGGCAUGAAUAAUGCCCACCUGUUGGUGCCC-UCGCCAUGGGCUCGUCCAUCUACUUA-------- ...((((.......((((((((((((((((((....))))))((((((....))))))........))))))...))))))((((-.......))))...........))))-------- ( -40.30) >DroYak_CAF1 37661 118 + 1 UUUUGAGGCACUUCGCCAGCUGGGGAGACCUUUGAUAGGGUCUCUGAAUUUAUUUGGCAUGAAUAAUGCCCACCUGUUGG-GCCC-UCGUAAUGGGCUCGUCCAUUUACUACCGUCUGCU .....((((......(((((.(((((((((((....)))))))))..........(((((.....)))))..)).)))))-((((-.......))))................))))... ( -41.00) >DroAna_CAF1 42435 102 + 1 GCUAAA-------------CCGGGGAGCUCUUUGAUAAGGUAAAAGACUUUUUUUCG-AUGAUUAAUUCACACCUGAGGUGGCCCAAUGCAAUGGGCCACGCUAUCUCCU----CUAACU ......-------------..(((((((((.((((.(((((.....)))))...)))-)(((.....))).....)))((((((((......)))))))).....)))))----)..... ( -31.50) >consensus CUUAAAGGCACCUCGCCAGCUGGGGAGACCUUUGAUAGGGUCGCCGAAUUUAUUCGGCAUGAGUAAUGCCCACCUGUUGGUGCCC_UCGCAAUGGGCUCGUCCAUCUACU____UGAGCU ..............((((((.((((.(((((......)))))((((((....))))))...........)).)).))))))((((........))))....................... (-23.27 = -25.22 + 1.95)

| Location | 17,169,963 – 17,170,078 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
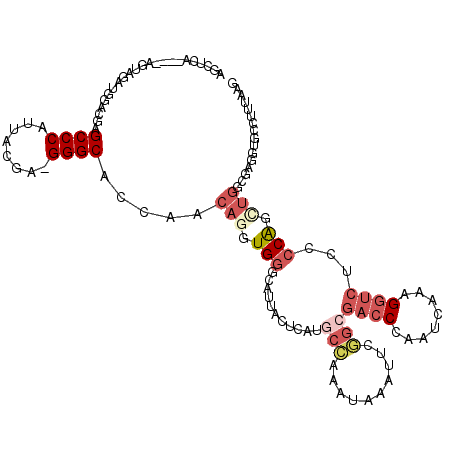
| Reading direction | reverse |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -36.68 |
| Consensus MFE | -14.28 |
| Energy contribution | -15.83 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.561095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17169963 115 - 20766785 AGCUCA----AGUCGAUGGACGAGCCCAUUGCGA-GGGCACCAACAGGUGGACAUUACUCAUGCCGAAUAAAUUCGGCGACCCAAUCAAAGGUCUCCCCAGAUGGCGAGAUGCCUUUAAG .((((.----.(.((((((......)))))))..-)))).......((.(((..........((((((....))))))((((........)))))))))....(((.....)))...... ( -36.40) >DroSec_CAF1 37124 117 - 1 AGCUCA--GUAGUAGAUGGACGAGCCCAUUAUGA-GGGCACCAACAGGUGGUUAUUACUCAUGCCGAAUAAAUUCGGCGACCCAAUCAAAGGUCUCCCCAGCUGGCAAGGUGCCUUUAAG ......--......(((((......))))).(((-(((((((..(((.(((...........((((((....))))))((((........))))...))).)))....)))))))))).. ( -42.90) >DroSim_CAF1 38191 115 - 1 AGCUCA----AGCAGAUGGACGAGCCCAUUAUGA-GGGCACCAACAGGUGGUUAUUACUCAUGCCGAAUAAAUUCGGCGACCCAAUCAAAGGUCUCCCCAGCUGGCAAGGUGCCUUUAAG .((...----.)).(((((......))))).(((-(((((((..(((.(((...........((((((....))))))((((........))))...))).)))....)))))))))).. ( -44.30) >DroEre_CAF1 38107 111 - 1 --------UAAGUAGAUGGACGAGCCCAUGGCGA-GGGCACCAACAGGUGGGCAUUAUUCAUGCCAAAUAAAUUCGGCGACCCUAUCAAAGGUCUGCCCAGUUGGCGAAGUGGCUUAAAA --------(((((..((...((.((((.......-)))).(((((...((((((........(((..........)))((((........)))))))))))))))))..)).)))))... ( -34.60) >DroYak_CAF1 37661 118 - 1 AGCAGACGGUAGUAAAUGGACGAGCCCAUUACGA-GGGC-CCAACAGGUGGGCAUUAUUCAUGCCAAAUAAAUUCAGAGACCCUAUCAAAGGUCUCCCCAGCUGGCGAAGUGCCUCAAAA (((....((((((((.(((......)))))))((-(.((-(((.....)))))....))).))))...........((((((........))))))....)))(((.....)))...... ( -35.10) >DroAna_CAF1 42435 102 - 1 AGUUAG----AGGAGAUAGCGUGGCCCAUUGCAUUGGGCCACCUCAGGUGUGAAUUAAUCAU-CGAAAAAAAGUCUUUUACCUUAUCAAAGAGCUCCCCGG-------------UUUAGC .(((((----(((((.....((((((((......))))))))(((.((((.........)))-)......(((........)))......)))))))....-------------)))))) ( -26.80) >consensus AGCUCA____AGUAGAUGGACGAGCCCAUUACGA_GGGCACCAACAGGUGGGCAUUACUCAUGCCAAAUAAAUUCGGCGACCCAAUCAAAGGUCUCCCCAGCUGGCGAGGUGCCUUUAAG .......................((((........)))).....(((.(((...........(((..........)))((((........))))...))).)))................ (-14.28 = -15.83 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:12 2006