| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,163,014 – 17,163,127 |
| Length | 113 |
| Max. P | 0.885469 |

| Location | 17,163,014 – 17,163,127 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.20 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -22.82 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
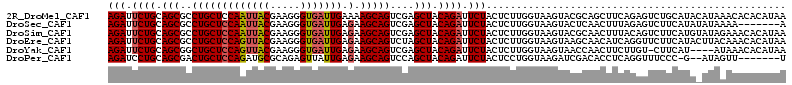
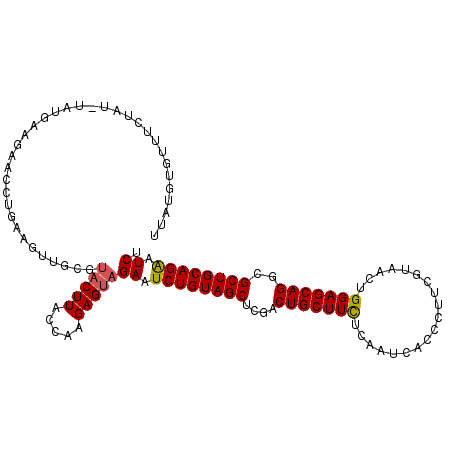
>2R_DroMel_CAF1 17163014 113 + 20766785 AGAUUCUGCAGCGCCUGCUCCAAUUACGAAGGGUGAUUGAAAAGCAGUCGAGCUACAGAUUCUACUCUUGGUAAGUACGCAGCUUCAGAGUCUGCAUACAUAAACACACAUAA (((.((((.(((..(((((.(((((((.....)))))))...)))))....))).)))).)))...........(((.((((((....)).)))).))).............. ( -32.00) >DroSec_CAF1 30203 106 + 1 AGAUUCUGCAGCGCCUGCUCCAAUUACGAAGGGUGAUUGAGAAGCAGUCGAGCUACAGAUUCUACUCUUGGUAAGUACUCAACUUUAGAGUCUUCAUAUAUAAAA-------A (((.((((.(((..(((((((((((((.....))))))).).)))))....))).)))).)))(((((....((((.....)))).)))))..............-------. ( -30.40) >DroSim_CAF1 31373 113 + 1 AGAUUCUGCAGCGCCUGCUCCAAUUACGAAGGGUGAUUGAGAAGCAGUCGAGCUACAGAUUCUACUCUUGGUAAGUACGCAACUUUACAGUCUUCAUGUAUAGAAACACAUAA (((.((((.(((..(((((((((((((.....))))))).).)))))....))).)))).)))...................((.((((.......)))).)).......... ( -28.00) >DroEre_CAF1 31151 113 + 1 AGAUUCUGCAGCGCCUGCUCCAGUUACGAAGGGUGAUUGAGAAGCAGUCUAGCUACAGAUUCUACUCUUGGUAAGUAAGCAACAUCAGGUUCUUCAUACUUACAAACACAUAA (((.((((.(((..(((((((((((((.....))))))).).)))))....))).)))).))).......(((((((((.(((.....)))))...))))))).......... ( -32.10) >DroYak_CAF1 30445 108 + 1 AGAUUCUGCAGCGGCUGCUCCAGUUACGAAGGGUGAUUGAGAAGCAGUCGAGCUACAGAUUCUACUCUUGGUAAGUAACCAACUUCUUGU-CUUCAU----AUAAACACAUAA (((.((((.((((((((((((((((((.....))))))).).)))))))..))).)))).)))....(((((.....)))))........-......----............ ( -31.60) >DroPer_CAF1 46408 103 + 1 AGAUCCUGCAGCGACUGCUCCAGAUGCGCAGAGUUAUUGAGAAGCAGUCCAGCUACAGAUUCUACUCCUGGUAAGAUCGACACCUCAGGUUUCCC-G--AUAGUU-------U (((..(((.((((((((((((((..((.....))..))).).)))))))..))).)))..)))(((..(((..(((((.........))))).))-)--..))).-------. ( -24.90) >consensus AGAUUCUGCAGCGCCUGCUCCAAUUACGAAGGGUGAUUGAGAAGCAGUCGAGCUACAGAUUCUACUCUUGGUAAGUACGCAACUUCAGAGUCUUCAUA_AUAAAAACACAUAA (((.((((.(((..(((((((((((((.....))))))).).)))))....))).)))).))).................................................. (-22.82 = -23.10 + 0.28)
| Location | 17,163,014 – 17,163,127 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.20 |
| Mean single sequence MFE | -32.63 |
| Consensus MFE | -23.74 |
| Energy contribution | -23.79 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
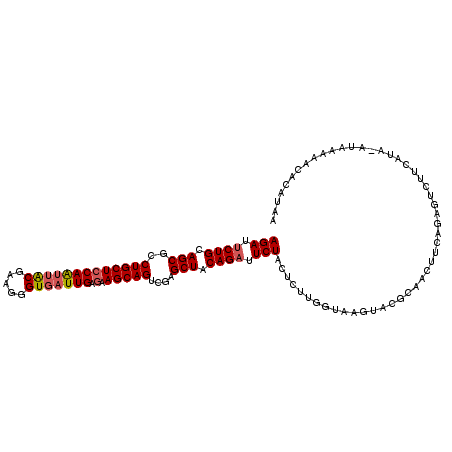
>2R_DroMel_CAF1 17163014 113 - 20766785 UUAUGUGUGUUUAUGUAUGCAGACUCUGAAGCUGCGUACUUACCAAGAGUAGAAUCUGUAGCUCGACUGCUUUUCAAUCACCCUUCGUAAUUGGAGCAGGCGCUGCAGAAUCU .(((.((.((....((((((((.((....))))))))))..))))...)))((.((((((((....((((((..((((.((.....)).))))))))))..)))))))).)). ( -36.40) >DroSec_CAF1 30203 106 - 1 U-------UUUUAUAUAUGAAGACUCUAAAGUUGAGUACUUACCAAGAGUAGAAUCUGUAGCUCGACUGCUUCUCAAUCACCCUUCGUAAUUGGAGCAGGCGCUGCAGAAUCU .-------..............(((((.((((.....))))....)))))(((.((((((((....(((((.(.((((.((.....)).))))))))))..)))))))).))) ( -29.00) >DroSim_CAF1 31373 113 - 1 UUAUGUGUUUCUAUACAUGAAGACUGUAAAGUUGCGUACUUACCAAGAGUAGAAUCUGUAGCUCGACUGCUUCUCAAUCACCCUUCGUAAUUGGAGCAGGCGCUGCAGAAUCU ((((((((....)))))))).((((....))))...(((((.....)))))((.((((((((....(((((.(.((((.((.....)).))))))))))..)))))))).)). ( -33.30) >DroEre_CAF1 31151 113 - 1 UUAUGUGUUUGUAAGUAUGAAGAACCUGAUGUUGCUUACUUACCAAGAGUAGAAUCUGUAGCUAGACUGCUUCUCAAUCACCCUUCGUAACUGGAGCAGGCGCUGCAGAAUCU ...((.((..(((((((...............)))))))..)))).....(((.((((((((....(((((((...................)))))))..)))))))).))) ( -31.77) >DroYak_CAF1 30445 108 - 1 UUAUGUGUUUAU----AUGAAG-ACAAGAAGUUGGUUACUUACCAAGAGUAGAAUCUGUAGCUCGACUGCUUCUCAAUCACCCUUCGUAACUGGAGCAGCCGCUGCAGAAUCU ...(((.(((..----..))).-))).....(((((.....)))))....(((.((((((((..(.(((((((...................)))))))).)))))))).))) ( -29.71) >DroPer_CAF1 46408 103 - 1 A-------AACUAU--C-GGGAAACCUGAGGUGUCGAUCUUACCAGGAGUAGAAUCUGUAGCUGGACUGCUUCUCAAUAACUCUGCGCAUCUGGAGCAGUCGCUGCAGGAUCU .-------.(((.(--(-((....))...((((.......)))).)))))(((.((((((((..(((((((((...................))))))))))))))))).))) ( -35.61) >consensus UUAUGUGUUUCUAU_UAUGAAGAACCUGAAGUUGCGUACUUACCAAGAGUAGAAUCUGUAGCUCGACUGCUUCUCAAUCACCCUUCGUAACUGGAGCAGGCGCUGCAGAAUCU ....................................(((((.....)))))((.((((((((....(((((((...................)))))))..)))))))).)). (-23.74 = -23.79 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:09 2006