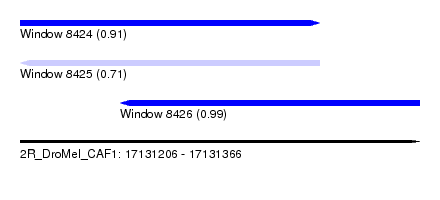
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,131,206 – 17,131,366 |
| Length | 160 |
| Max. P | 0.994468 |

| Location | 17,131,206 – 17,131,326 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -45.22 |
| Consensus MFE | -39.74 |
| Energy contribution | -39.82 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17131206 120 + 20766785 AGGAUAGUAGUUUCGGGGAGUAGCUUAGGCGACUGGCCUCCAGUCCCGGCGUCGUGGAGGACUCAUCCUUAAACAGGAUGGUGGGUGAGUUGUGCUCGCUCUUCGACAAGUACAAGCUGU ......(((.((((((((((((((....((((((.(((..(((((((.((...)).).)))))((((((.....)))))))..))).))))))))).))))))))).)).)))....... ( -46.80) >DroSec_CAF1 67094 120 + 1 AGGAUAGUAGUUUCGGGGAGUAGCUUAGGCGACUGGCCUCCAGUCCCGGCGUCGUGGAGGACUCAUCCUUAAACAGGAUGGUGGGUGAGUUGUGCUCGCUCUUAGACAAGUACAAGCUGU ......(((.((((..((((((((....((((((.(((..(((((((.((...)).).)))))((((((.....)))))))..))).))))))))).)))))..)).)).)))....... ( -42.40) >DroSim_CAF1 57939 120 + 1 AGGAUAGUAGUUUCGGGGAGUAGCUUAGGCGACUGGCCUCCAGUCCCGGCGUCGUGGAGGACUCAUCCUUAAACAGGAUGGUGGGUGAGUUGUGCUCGCUCUUCGACAAGUACAAGCUGU ......(((.((((((((((((((....((((((.(((..(((((((.((...)).).)))))((((((.....)))))))..))).))))))))).))))))))).)).)))....... ( -46.80) >DroEre_CAF1 72616 120 + 1 AGGAUAGCAGUUUCGGGGAGUAGCUCAGGCGACUGGCCUCCAGUCCCGGCGUCGUAGAGGACUCAUCCUUAAACAGGAUGGUCGGUGAGUUAUGCUCGCUCUUUGACAAGUACAAGCUGU ......((((((((((((.(..((.(((....)))))..)...)))))..((((.(((.(((.((((((.....))))))))).(((((.....)))))))).))))......))))))) ( -45.10) >DroYak_CAF1 74513 120 + 1 AGGACAACAGUUUUGGGGAGUAACUCAGGCGACUGGCCUCCAGUCCCGGCGUCGUCGAGGACUCAUCCUUAAACAGGAUGGUGGGUGAGUUGUGCUCGCUCUUCGACAAGUACAAGCUGU ......(((((((((((......))))((.((((((...))))))))......(((((((((.((((((.....)))))))).((((((.....)))))))))))))......))))))) ( -45.00) >consensus AGGAUAGUAGUUUCGGGGAGUAGCUUAGGCGACUGGCCUCCAGUCCCGGCGUCGUGGAGGACUCAUCCUUAAACAGGAUGGUGGGUGAGUUGUGCUCGCUCUUCGACAAGUACAAGCUGU .......(((((((((((((((((....((((((.(((..(((((((.((...)).).)))))((((((.....)))))))..))).))))))))).)))))))))........))))). (-39.74 = -39.82 + 0.08)

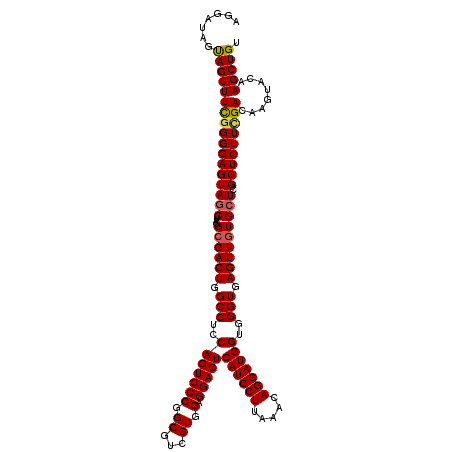
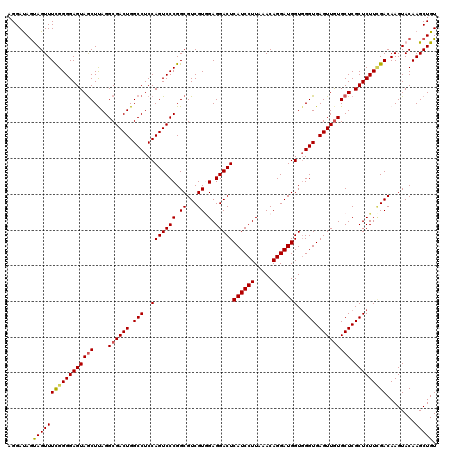
| Location | 17,131,206 – 17,131,326 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -35.06 |
| Consensus MFE | -32.38 |
| Energy contribution | -32.38 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17131206 120 - 20766785 ACAGCUUGUACUUGUCGAAGAGCGAGCACAACUCACCCACCAUCCUGUUUAAGGAUGAGUCCUCCACGACGCCGGGACUGGAGGCCAGUCGCCUAAGCUACUCCCCGAAACUACUAUCCU ..((((((.....((((..(((.(((.....)))....((((((((.....)))))).)).)))..))))...((((((((...)))))).))))))))..................... ( -34.70) >DroSec_CAF1 67094 120 - 1 ACAGCUUGUACUUGUCUAAGAGCGAGCACAACUCACCCACCAUCCUGUUUAAGGAUGAGUCCUCCACGACGCCGGGACUGGAGGCCAGUCGCCUAAGCUACUCCCCGAAACUACUAUCCU ...((((((.(((....))).)))))).............((((((.....)))))).(((......)))..(((((((((...))))))((....))......)))............. ( -31.80) >DroSim_CAF1 57939 120 - 1 ACAGCUUGUACUUGUCGAAGAGCGAGCACAACUCACCCACCAUCCUGUUUAAGGAUGAGUCCUCCACGACGCCGGGACUGGAGGCCAGUCGCCUAAGCUACUCCCCGAAACUACUAUCCU ..((((((.....((((..(((.(((.....)))....((((((((.....)))))).)).)))..))))...((((((((...)))))).))))))))..................... ( -34.70) >DroEre_CAF1 72616 120 - 1 ACAGCUUGUACUUGUCAAAGAGCGAGCAUAACUCACCGACCAUCCUGUUUAAGGAUGAGUCCUCUACGACGCCGGGACUGGAGGCCAGUCGCCUGAGCUACUCCCCGAAACUGCUAUCCU ..(((((......(((..((((.(((.....)))...(((((((((.....)))))).)))))))..)))...((((((((...)))))).)).)))))..................... ( -36.50) >DroYak_CAF1 74513 120 - 1 ACAGCUUGUACUUGUCGAAGAGCGAGCACAACUCACCCACCAUCCUGUUUAAGGAUGAGUCCUCGACGACGCCGGGACUGGAGGCCAGUCGCCUGAGUUACUCCCCAAAACUGUUGUCCU ...((((((.(((....))).))))))..(((((..((..((((((.....))))))(((((.((.......)))))))))((((.....)))))))))..................... ( -37.60) >consensus ACAGCUUGUACUUGUCGAAGAGCGAGCACAACUCACCCACCAUCCUGUUUAAGGAUGAGUCCUCCACGACGCCGGGACUGGAGGCCAGUCGCCUAAGCUACUCCCCGAAACUACUAUCCU ..((((((.....((((..(((.(((.....)))....((((((((.....)))))).)).)))..))))...((((((((...)))))).))))))))..................... (-32.38 = -32.38 + -0.00)

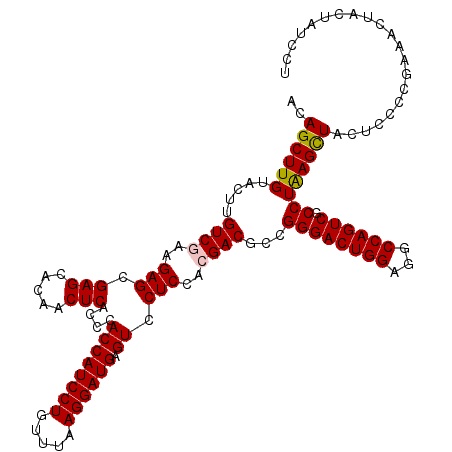
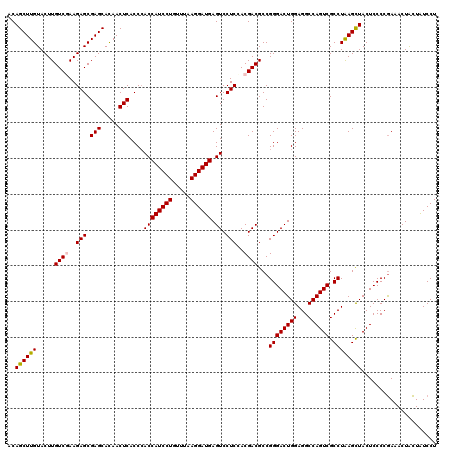
| Location | 17,131,246 – 17,131,366 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.92 |
| Mean single sequence MFE | -42.26 |
| Consensus MFE | -38.02 |
| Energy contribution | -37.38 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
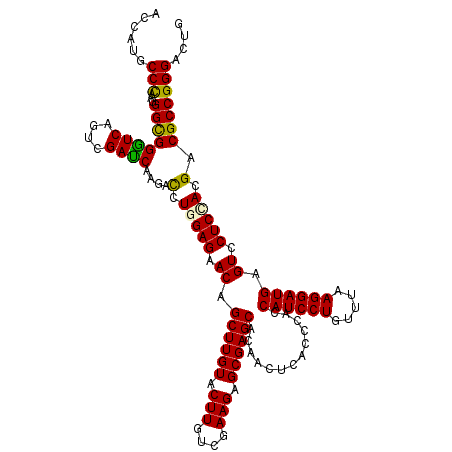
>2R_DroMel_CAF1 17131246 120 - 20766785 ACUAUGCCCAAGGGCGGGUCAGUCGACCAAGACCUAGAGAACAGCUUGUACUUGUCGAAGAGCGAGCACAACUCACCCACCAUCCUGUUUAAGGAUGAGUCCUCCACGACGCCGGGACUG .....(((....)))((((..(((......)))...(((....((((((.(((....))).))))))....)))))))..((((((.....))))))((((((..........)))))). ( -40.50) >DroSec_CAF1 67134 120 - 1 ACCAUGCCCAAGGGCGGGUCAGUCGAUCAAGGCCUGGAGAACAGCUUGUACUUGUCUAAGAGCGAGCACAACUCACCCACCAUCCUGUUUAAGGAUGAGUCCUCCACGACGCCGGGACUG (((.((((....)))))))(((((......(((.(((((.((.((((((.(((....))).)))))).............((((((.....)))))).)).)))))....)))..))))) ( -42.80) >DroSim_CAF1 57979 120 - 1 ACCAUGCCCAAGGGCGGGUCAGUCGAUCAAGACAUGGAGAACAGCUUGUACUUGUCGAAGAGCGAGCACAACUCACCCACCAUCCUGUUUAAGGAUGAGUCCUCCACGACGCCGGGACUG .((..(((....)))(((((.(((......))).(((((.((.((((((.(((....))).)))))).............((((((.....)))))).)).))))).))).))))..... ( -44.70) >DroEre_CAF1 72656 120 - 1 ACCAUGCCUAAGGGCGGAUCUGUCGAUCAAGAUCUGGAGAACAGCUUGUACUUGUCAAAGAGCGAGCAUAACUCACCGACCAUCCUGUUUAAGGAUGAGUCCUCUACGACGCCGGGACUG ......(((...(((((((((........)))))(((((....((((((.(((....))).))))))..........(((((((((.....)))))).))))))))...))))))).... ( -41.10) >DroYak_CAF1 74553 120 - 1 ACCAUACCUAAGGGUGGAUCGGUCGAUCAAGACCUGGAGAACAGCUUGUACUUGUCGAAGAGCGAGCACAACUCACCCACCAUCCUGUUUAAGGAUGAGUCCUCGACGACGCCGGGACUG ...........((((((...((((......)))).........((((((.(((....))).)))))).....))))))..((((((.....))))))(((((.((.......))))))). ( -42.20) >consensus ACCAUGCCCAAGGGCGGGUCAGUCGAUCAAGACCUGGAGAACAGCUUGUACUUGUCGAAGAGCGAGCACAACUCACCCACCAUCCUGUUUAAGGAUGAGUCCUCCACGACGCCGGGACUG ......(((...((((((((....))))....(.(((((.((.((((((.(((....))).)))))).............((((((.....)))))).)).))))).).))))))).... (-38.02 = -37.38 + -0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:50 2006