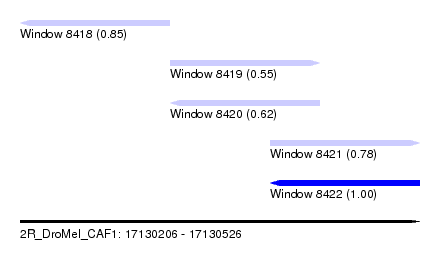
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,130,206 – 17,130,526 |
| Length | 320 |
| Max. P | 0.999853 |

| Location | 17,130,206 – 17,130,326 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -31.28 |
| Energy contribution | -31.68 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17130206 120 - 20766785 UGGUCCUAAGGAAUCGCCUUUCAAGCAGAAGUCAAUGGAUUUGCCGAUGCCGACACUCCAGGCAAAGACCACAGUGUCUACAUCAUCGAUGAAUCUCCUUCAGCGUCGGGGAUCCAAUCA (((((((((((.....))))....((.((((.....((((((..(((((..((((((...((......))..)))))).....)))))..)))))).)))).))....)))).))).... ( -33.40) >DroSec_CAF1 66097 120 - 1 UGGACCUAAGGAAUCGCCUUUCAAGCAGAAGUCAAUGGAUCUGCCGAUGCCGACACUCCAGGCAAAGAACACAGUGUCUACAUCAUCGAUGAAUCUCCUUCAGCGUCGGGGAUCCAACCA .((....((((.....))))...............(((((((.(((((((.((((((...(........)..))))))..((((...))))...........)))))))))))))).)). ( -34.00) >DroSim_CAF1 56942 120 - 1 UGGUCCUAAGGAAUCGCCUUUCAAGCAGAAGUCAAUGGAUCUGCCGAUGCCGACACUCCAGGCAAAGACCACAGUGUCUACAUCAUCGAUGAAUCUCCUUCAGCGUCGGGGAUCCAACCA .(((...((((.....))))...............(((((((.(((((((.((((((...((......))..))))))..((((...))))...........))))))))))))))))). ( -36.70) >DroEre_CAF1 71619 120 - 1 UGGUCCUAAGGAAUCGCCUUUCAAGCAGAAGUCAAUGGAUCUGCCGAUGCCGACACUGCAGGCAAAGACCACAGUGUCUACAUCAUCUAUGAAUCUCCUCCAACGUCGGGGAUCCAAUCA ..((...((((.....))))....)).........(((((((.((((((..(((((((..((......)).)))))))....(((....)))...........))))))))))))).... ( -33.00) >DroYak_CAF1 73516 120 - 1 UGGUGCUAAGGAAUCGCCUUUCAAGCAGAAGUCUAUGGAUCUGCCGAUGCCGACACUCCAGGCAAAGACCACAGUGUCCACAUCAUCGAUGAAUCUUCUCCAGCGUCGGGGAUCCAAUCA ...((((((((.....))))...))))........(((((((.(((((((.((((((...((......))..))))))..((((...))))...........)))))))))))))).... ( -37.90) >consensus UGGUCCUAAGGAAUCGCCUUUCAAGCAGAAGUCAAUGGAUCUGCCGAUGCCGACACUCCAGGCAAAGACCACAGUGUCUACAUCAUCGAUGAAUCUCCUUCAGCGUCGGGGAUCCAAUCA .(((...((((.....))))...............(((((((.(((((((.((((((...((......))..))))))..((((...))))...........))))))))))))))))). (-31.28 = -31.68 + 0.40)

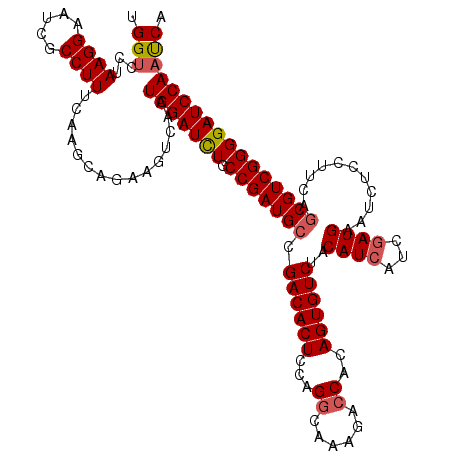
| Location | 17,130,326 – 17,130,446 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -46.04 |
| Consensus MFE | -43.48 |
| Energy contribution | -44.12 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17130326 120 + 20766785 CCGCCAGAGGAGGAGAACUCCACCAGGGUAAGGCACUCGGGAAGCGUUUUUAAAUUUGUGCUGGACGAGUGCAGUAACUGAUUGUGCUUGUGGGCGCUUCUCGGCGCUAGGCUUGCGCCU ..(((...((((.....))))((....))..)))....(((((((((((...............(((((..((((.....))))..))))))))))))))))(((((.......))))). ( -47.56) >DroSec_CAF1 66217 120 + 1 CCGCCAGAGGAGGAGAACUCCACCAGGGUAAGGCACUCCGGAAGCGUUUUUAAAUUUGUGCUGGACGAGUGCAGUAACUGAUUGUGCUUGUGGGCGCUUCUCGGCGCUAGGCUUGCACCU ........((((.....))))...((((((((.(.(.((((((((((((...............(((((..((((.....))))..)))))))))))))).))).)...).))))).))) ( -41.66) >DroSim_CAF1 57062 120 + 1 CCGCCAGAGGAGGAGAACUCCACCAGGGUAAGGCACUCCGGAAGCGUUUUUAAAUUUGUGCUGGACGAGUGCAGUAACUGAUUGUGCUUGUGGGCGCUUCUCGGCGCUAGGCUUGCACCU ........((((.....))))...((((((((.(.(.((((((((((((...............(((((..((((.....))))..)))))))))))))).))).)...).))))).))) ( -41.66) >DroEre_CAF1 71739 120 + 1 CCGCCAGUGGAGGAGAACUCCACCAGGGUAAGGCACUCGGGAAGCGUCUUUAAAUUUGUGCUGGACGAGUGCAGUAACUGAUUGUGCUUGUGGGCGCUUCUCGGCGCUAGGCUUGCGCCU ..(((.((((((.....))))))...)))..(((.(.((((((((((((...............(((((..((((.....))))..)))))))))))))))))).)))((((....)))) ( -51.76) >DroYak_CAF1 73636 120 + 1 CCGCCAGAGGAGGAGAACUCCACCAGGGUAAGGCACUCGGGAAGCGUUUUUAAAUUUGUGCUGGACGAGUGCAGUAACUGAUUGUGCUUGUGGGCGCUUCUCGGCGCUAGGCUUGCGCCU ..(((...((((.....))))((....))..)))....(((((((((((...............(((((..((((.....))))..))))))))))))))))(((((.......))))). ( -47.56) >consensus CCGCCAGAGGAGGAGAACUCCACCAGGGUAAGGCACUCGGGAAGCGUUUUUAAAUUUGUGCUGGACGAGUGCAGUAACUGAUUGUGCUUGUGGGCGCUUCUCGGCGCUAGGCUUGCGCCU ..(((...((((.....))))((....))..)))....(((((((((((...............(((((..((((.....))))..))))))))))))))))(((((.......))))). (-43.48 = -44.12 + 0.64)

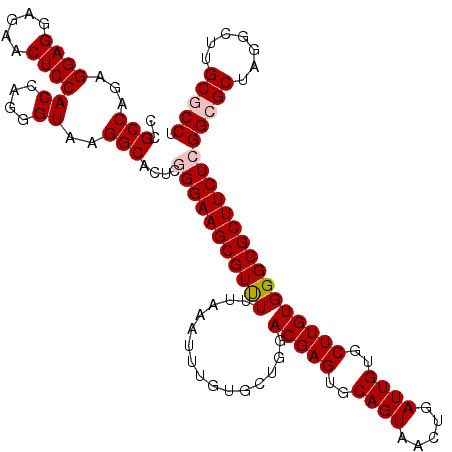
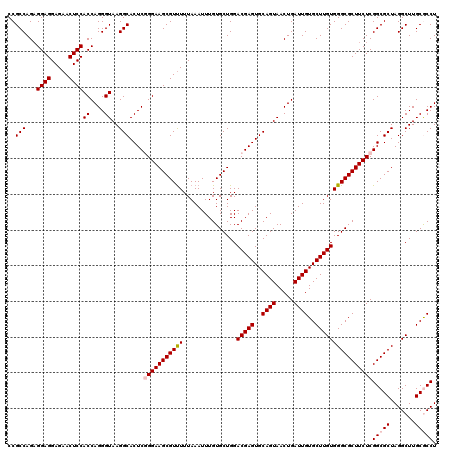
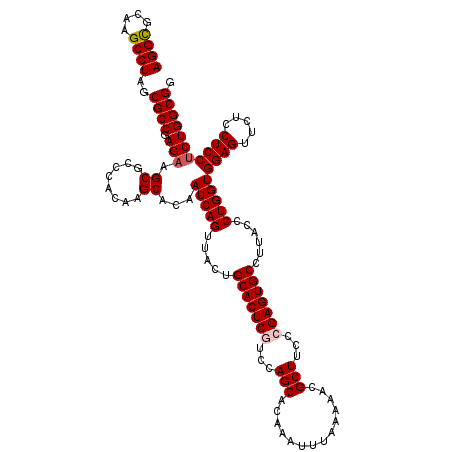
| Location | 17,130,326 – 17,130,446 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -35.88 |
| Energy contribution | -36.24 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17130326 120 - 20766785 AGGCGCAAGCCUAGCGCCGAGAAGCGCCCACAAGCACAAUCAGUUACUGCACUCGUCCAGCACAAAUUUAAAAACGCUUCCCGAGUGCCUUACCCUGGUGGAGUUCUCCUCCUCUGGCGG ((((....))))..((((.(((.((........))...(((((.....(((((((...(((..............)))...)))))))......)))))((((.....))))))))))). ( -40.24) >DroSec_CAF1 66217 120 - 1 AGGUGCAAGCCUAGCGCCGAGAAGCGCCCACAAGCACAAUCAGUUACUGCACUCGUCCAGCACAAAUUUAAAAACGCUUCCGGAGUGCCUUACCCUGGUGGAGUUCUCCUCCUCUGGCGG .(((((.......)))))......((((..........(((((.....((((((....(((..............)))....))))))......)))))((((.....))))...)))). ( -33.14) >DroSim_CAF1 57062 120 - 1 AGGUGCAAGCCUAGCGCCGAGAAGCGCCCACAAGCACAAUCAGUUACUGCACUCGUCCAGCACAAAUUUAAAAACGCUUCCGGAGUGCCUUACCCUGGUGGAGUUCUCCUCCUCUGGCGG .(((((.......)))))......((((..........(((((.....((((((....(((..............)))....))))))......)))))((((.....))))...)))). ( -33.14) >DroEre_CAF1 71739 120 - 1 AGGCGCAAGCCUAGCGCCGAGAAGCGCCCACAAGCACAAUCAGUUACUGCACUCGUCCAGCACAAAUUUAAAGACGCUUCCCGAGUGCCUUACCCUGGUGGAGUUCUCCUCCACUGGCGG ((((....)))).((((......)))).....................(((((((...(((..............)))...)))))))....(((..((((((.....))))))..).)) ( -42.24) >DroYak_CAF1 73636 120 - 1 AGGCGCAAGCCUAGCGCCGAGAAGCGCCCACAAGCACAAUCAGUUACUGCACUCGUCCAGCACAAAUUUAAAAACGCUUCCCGAGUGCCUUACCCUGGUGGAGUUCUCCUCCUCUGGCGG ((((....))))..((((.(((.((........))...(((((.....(((((((...(((..............)))...)))))))......)))))((((.....))))))))))). ( -40.24) >consensus AGGCGCAAGCCUAGCGCCGAGAAGCGCCCACAAGCACAAUCAGUUACUGCACUCGUCCAGCACAAAUUUAAAAACGCUUCCCGAGUGCCUUACCCUGGUGGAGUUCUCCUCCUCUGGCGG ((((....))))..((((.(((.((........))...(((((.....(((((((...(((..............)))...)))))))......)))))((((.....))))))))))). (-35.88 = -36.24 + 0.36)

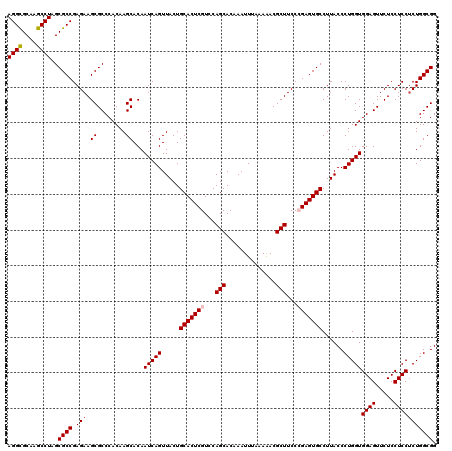
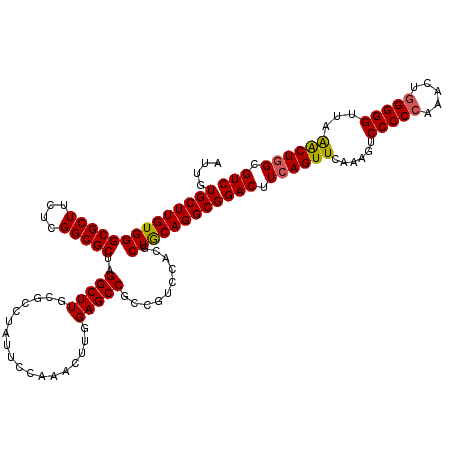
| Location | 17,130,406 – 17,130,526 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -46.76 |
| Consensus MFE | -40.21 |
| Energy contribution | -40.05 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17130406 120 + 20766785 AUUGUGCUUGUGGGCGCUUCUCGGCGCUAGGCUUGCGCCUAUUCCAAACUUCGAGCCACCGUCCACGCUACAGGCGGACUUCAGUUCAAAGUCCCCCAAACUGGGGGUUAAACUGGCGUC ....((((((((((((((....)))))..((((((................))))))..........)))))))))(((.((((((......(((((.....)))))...)))))).))) ( -43.99) >DroSec_CAF1 66297 120 + 1 AUUGUGCUUGUGGGCGCUUCUCGGCGCUAGGCUUGCACCUAUUCCAAACUUUGAGCCACCGUCCACGCUGCAGGCGGACUUCAGUUCAAAGUCCCCCAAACUGGGGGUUAAACUGGCGUC ...((((..((.((((((....))))))..))..)))).....(((.(((((((((....((((..((.....))))))....)))))))))(((((.....)))))......))).... ( -44.80) >DroSim_CAF1 57142 120 + 1 AUUGUGCUUGUGGGCGCUUCUCGGCGCUAGGCUUGCACCUAUUCCAAACUUUGAGCCGCCGUCCACGCUGCAGGCGGACUUCAGUUCAAAGUCCCCCAAACUGGGGGUUAAACUGGCGUC ...((((..((.((((((....))))))..))..)))).....(((.(((((((((.(..((((..((.....))))))..).)))))))))(((((.....)))))......))).... ( -45.50) >DroEre_CAF1 71819 120 + 1 AUUGUGCUUGUGGGCGCUUCUCGGCGCUAGGCUUGCGCCUAUUCCAAACUUGGAGCCGCCGUCCACGCUGCAGGCGGACUUCAGUUCAAAGUCCCCCAGGCUGGGGGUUAAGCUGGCGUC ....((((((..(.((.....(((((.(((((....)))))(((((....))))).)))))....)))..))))))(((.((((((......(((((.....)))))...)))))).))) ( -49.20) >DroYak_CAF1 73716 120 + 1 AUUGUGCUUGUGGGCGCUUCUCGGCGCUAGGCUUGCGCCUAUUCCAAACUUGGAGCCGCCGUCCACGCUGCAGGCGGACUUCAGUUCAAAGUCCCCAAGGCUGGGGGUUAGGCUUGCGUC .....((..(((((((.....((((..(((((....))))).((((....)))))))).))))))))).((((((((((....))))..(..(((((....)))))..)..))))))... ( -50.30) >consensus AUUGUGCUUGUGGGCGCUUCUCGGCGCUAGGCUUGCGCCUAUUCCAAACUUGGAGCCGCCGUCCACGCUGCAGGCGGACUUCAGUUCAAAGUCCCCCAAACUGGGGGUUAAACUGGCGUC ....((((((((((((((....)))))..(((((..................)))))..........)))))))))(((.((((((......(((((.....)))))...)))))).))) (-40.21 = -40.05 + -0.16)

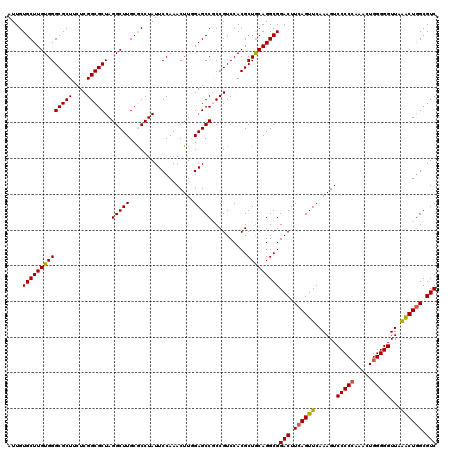
| Location | 17,130,406 – 17,130,526 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -48.66 |
| Consensus MFE | -46.96 |
| Energy contribution | -46.96 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.26 |
| SVM RNA-class probability | 0.999853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
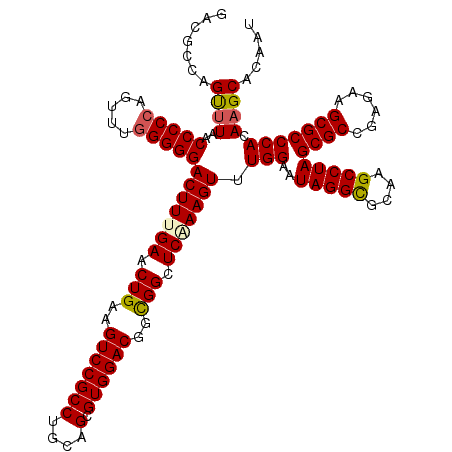
>2R_DroMel_CAF1 17130406 120 - 20766785 GACGCCAGUUUAACCCCCAGUUUGGGGGACUUUGAACUGAAGUCCGCCUGUAGCGUGGACGGUGGCUCGAAGUUUGGAAUAGGCGCAAGCCUAGCGCCGAGAAGCGCCCACAAGCACAAU ...(((((((((((((((.....)))))...))))))))..(((((((....).)))))).((((..((...(((((..(((((....)))))...)))))...)).))))..))..... ( -52.80) >DroSec_CAF1 66297 120 - 1 GACGCCAGUUUAACCCCCAGUUUGGGGGACUUUGAACUGAAGUCCGCCUGCAGCGUGGACGGUGGCUCAAAGUUUGGAAUAGGUGCAAGCCUAGCGCCGAGAAGCGCCCACAAGCACAAU ..((((((((((((((((.....)))))...)))))))...(((((((....).)))))))))).......(((((...(((((....)))))((((......))))...)))))..... ( -46.30) >DroSim_CAF1 57142 120 - 1 GACGCCAGUUUAACCCCCAGUUUGGGGGACUUUGAACUGAAGUCCGCCUGCAGCGUGGACGGCGGCUCAAAGUUUGGAAUAGGUGCAAGCCUAGCGCCGAGAAGCGCCCACAAGCACAAU ..((((((((((((((((.....)))))...)))))))...(((((((....).)))))))))).......(((((...(((((....)))))((((......))))...)))))..... ( -48.20) >DroEre_CAF1 71819 120 - 1 GACGCCAGCUUAACCCCCAGCCUGGGGGACUUUGAACUGAAGUCCGCCUGCAGCGUGGACGGCGGCUCCAAGUUUGGAAUAGGCGCAAGCCUAGCGCCGAGAAGCGCCCACAAGCACAAU .......((((..(((((.....)))))((((.((.(((..(((((((....).))))))..))).)).)))).(((..(((((....)))))((((......))))))).))))..... ( -50.10) >DroYak_CAF1 73716 120 - 1 GACGCAAGCCUAACCCCCAGCCUUGGGGACUUUGAACUGAAGUCCGCCUGCAGCGUGGACGGCGGCUCCAAGUUUGGAAUAGGCGCAAGCCUAGCGCCGAGAAGCGCCCACAAGCACAAU ...((..(((....((((......)))).............(((((((....).)))))))))(((.(....(((((..(((((....)))))...)))))..).))).....))..... ( -45.90) >consensus GACGCCAGUUUAACCCCCAGUUUGGGGGACUUUGAACUGAAGUCCGCCUGCAGCGUGGACGGCGGCUCAAAGUUUGGAAUAGGCGCAAGCCUAGCGCCGAGAAGCGCCCACAAGCACAAU .......((((..(((((.....)))))(((((((.(((..(((((((....).))))))..))).))))))).(((..(((((....)))))((((......))))))).))))..... (-46.96 = -46.96 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:46 2006