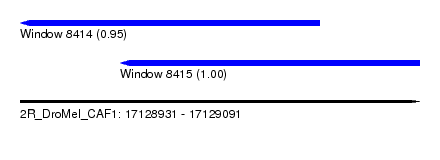
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,128,931 – 17,129,091 |
| Length | 160 |
| Max. P | 0.996902 |

| Location | 17,128,931 – 17,129,051 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -42.08 |
| Consensus MFE | -37.34 |
| Energy contribution | -37.42 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17128931 120 - 20766785 UAUCUUGCCAAACGAGAACUCUCGCGUGCUACUCGAAUCCGAGUCCAGCGAGCUGACUAGCUUGCUGGGGGAAAUAAAACGGACCAGCUCCGUGACCGCUAGCGAGGACUUGCCCUACAU ..(((((.....)))))..((((((..((.(((((....)))))((((((((((....))))))))))((........(((((.....)))))..))))..))))))............. ( -42.00) >DroSec_CAF1 64822 120 - 1 UAUCUUGCCAAAUGAGAACUCUCGCGUUCUACUCGAAUCCGAGUCCAGCGAGCUGAUUAGCUUGCUGGGGGAAGUAAAACGGACCAGCUCCGUGACCGCCAGCGAGGACUUGCCCUACAU ..((((((.....(((....)))((((((((((((....)))))((((((((((....)))))))))).))))((...(((((.....))))).)))))..))))))............. ( -42.70) >DroSim_CAF1 55667 120 - 1 UAUCUUGCCAAAUGAGAACUCUCGCGUUCUACUCGAAUCCGAGUCCAGCGAGCUGAUUAGCUUGCUGGGGGAAGUAAAACGGACCAGCUCCGUGACCGCCAGUGAGGACUUGCCCUACAU ..............(((((......)))))(((((....)))))((((((((((....))))))))))(((((((...(((((.....)))))..((........)))))).)))..... ( -40.70) >DroEre_CAF1 70347 120 - 1 UAUCUUGCCCAACGAGAACUCUCGCGUGCUUCUCGAAUCCGAGUCCAGCGAGCUGACUAGCUUGCUGGGAGAAGUCAAACGGACCAGCUCCGUGACCGCCAGCGAGGAUUUGCCCUACAU ..((((((.....(((....)))(((.((((((((....)....((((((((((....)))))))))))))))))...(((((.....)))))...)))..))))))............. ( -42.70) >DroYak_CAF1 72241 120 - 1 UAUCUUGCCCAACGAGAACUCUCGCGUGCUGCUCGAAUCCGAGUCCAGCGAGCUGACUCGCUUGCUGGGGGAAGUCAAACGAACCAGUUCCGUGACCGCCAGCGAGGACUUGCCCUACAU ..(((((.....)))))..((((((.....(((((....)))))(((((((((......))))))))).((..((((...(((....)))..))))..)).))))))............. ( -42.30) >consensus UAUCUUGCCAAACGAGAACUCUCGCGUGCUACUCGAAUCCGAGUCCAGCGAGCUGACUAGCUUGCUGGGGGAAGUAAAACGGACCAGCUCCGUGACCGCCAGCGAGGACUUGCCCUACAU ..(((((.....)))))..((((((......((((....))))((((((((((......))))))))))((..((...(((((.....))))).))..)).))))))............. (-37.34 = -37.42 + 0.08)

| Location | 17,128,971 – 17,129,091 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -38.86 |
| Consensus MFE | -34.49 |
| Energy contribution | -34.57 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17128971 120 - 20766785 CAAUGGUCUUCGGCUCGCAGACAAAAAAUCGCUACAAGACUAUCUUGCCAAACGAGAACUCUCGCGUGCUACUCGAAUCCGAGUCCAGCGAGCUGACUAGCUUGCUGGGGGAAAUAAAAC ......((((((((.(((................((((.....))))......(((....)))))).)))(((((....))))).(((((((((....))))))))))))))........ ( -37.60) >DroSec_CAF1 64862 120 - 1 CCAUGGUCUUCGGCUCGCAGACAAAAAAUCGCUACAAGACUAUCUUGCCAAAUGAGAACUCUCGCGUUCUACUCGAAUCCGAGUCCAGCGAGCUGAUUAGCUUGCUGGGGGAAGUAAAAC ..((((((((.(((................)))..))))))))(((.((.....(((((......)))))(((((....)))))((((((((((....)))))))))))).)))...... ( -38.79) >DroSim_CAF1 55707 120 - 1 CCAUGGUCUUCGGCUCGCAGACAAAAAAUCGCUACAAGACUAUCUUGCCAAAUGAGAACUCUCGCGUUCUACUCGAAUCCGAGUCCAGCGAGCUGAUUAGCUUGCUGGGGGAAGUAAAAC ..((((((((.(((................)))..))))))))(((.((.....(((((......)))))(((((....)))))((((((((((....)))))))))))).)))...... ( -38.79) >DroEre_CAF1 70387 120 - 1 CUAUGGUUUUCGGCUCACAGACGAAAAAUCGCUACAAGACUAUCUUGCCCAACGAGAACUCUCGCGUGCUUCUCGAAUCCGAGUCCAGCGAGCUGACUAGCUUGCUGGGAGAAGUCAAAC .....(((((((......((.(((....))))).((((.....)))).....)))))))((((........((((....)))).((((((((((....))))))))))))))........ ( -37.50) >DroYak_CAF1 72281 120 - 1 CUAUGGUCUUCGGCUCGCAGACAAAAAAUCGCUACAAGACUAUCUUGCCCAACGAGAACUCUCGCGUGCUGCUCGAAUCCGAGUCCAGCGAGCUGACUCGCUUGCUGGGGGAAGUCAAAC ...(((.(((((((.(((................((((.....))))......(((....)))))).)))(((((....)))))(((((((((......)))))))))..)))))))... ( -41.60) >consensus CCAUGGUCUUCGGCUCGCAGACAAAAAAUCGCUACAAGACUAUCUUGCCAAACGAGAACUCUCGCGUGCUACUCGAAUCCGAGUCCAGCGAGCUGACUAGCUUGCUGGGGGAAGUAAAAC ..((((((((.(((................)))..))))))))(((.((...((((....))))......(((((....)))))(((((((((......))))))))))).)))...... (-34.49 = -34.57 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:38 2006