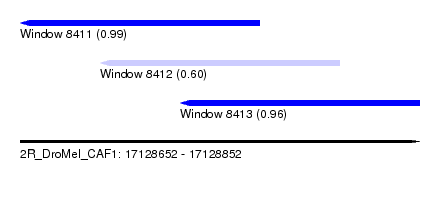
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,128,652 – 17,128,852 |
| Length | 200 |
| Max. P | 0.991123 |

| Location | 17,128,652 – 17,128,772 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -40.92 |
| Consensus MFE | -36.02 |
| Energy contribution | -36.46 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17128652 120 - 20766785 CCAGAACACGCAGCGCUACAUAAGACGGUGCGUGGAUGGAGGCAGCAGCAGCAGUCCUCACGUUGACAGGGAGCAAAUCCUGCAGCAGUACUUCCAGAAGAUUGUCAUGCUCACAAACUU (((...((((((.((((.....)).)).))))))..))).(..((((...((((((.((..((((.(((((......))))))))).(......).)).))))))..))))..)...... ( -39.00) >DroSec_CAF1 64545 120 - 1 CCAGAACACGCAGCGCUACAUAAGACGGUGCGUGGAUGGGGGCAGCAGCAGCAGUCCUCACGUUGACAGGGAGCAAAUCCUGCAGCAGUACUUCCAGAAGAUUGUCAUGCUCACAAACUU (((...((((((.((((.....)).)).))))))..)))(((((......((((((.((..((((.(((((......))))))))).(......).)).))))))..)))))........ ( -39.60) >DroSim_CAF1 55390 120 - 1 CCAGAACACGCAGCGCUACAUAAGACGGUGCGUGGAUGGGGGCAGCAGCAGCAGUCCUCACGUUGACAGGGAGCAAAUCCUGCAGCAGUACUUCCAGAAGAUUGUCAUGCUCACAAACUU (((...((((((.((((.....)).)).))))))..)))(((((......((((((.((..((((.(((((......))))))))).(......).)).))))))..)))))........ ( -39.60) >DroEre_CAF1 70073 120 - 1 CCAGAACACGCAGCGUUACAUUCGACGCUGCGUUGAUGGUGGAAGCAGCAGCAGUCCUCACGUUGACAGGGAGCAAAUCCUGCAGCAGUACUUCCAGAAGAUUGUCAUGCUCACCAACUU .......((((((((((......))))))))))...(((((..((((...((((((.((..((((.(((((......))))))))).(......).)).))))))..))))))))).... ( -43.60) >DroYak_CAF1 71967 120 - 1 CCAAAACACGCAGCGCUACAUACGUCGCUGCGUGGAUGGAGGAAGCAGCAGCAGUCCUCACGUUGACAGGGAGCAAAUCCUGCAGCAGUACUUUCAGAAGAUUGUCAUGCUCACAAACUU (((...((((((((((.......).)))))))))..))).(..((((...((((((.((..((((.(((((......))))))))).(......).)).))))))..))))..)...... ( -42.80) >consensus CCAGAACACGCAGCGCUACAUAAGACGGUGCGUGGAUGGAGGCAGCAGCAGCAGUCCUCACGUUGACAGGGAGCAAAUCCUGCAGCAGUACUUCCAGAAGAUUGUCAUGCUCACAAACUU (((...((((((.((((.....)).)).))))))..))).(..((((...((((((.((..((((.(((((......))))))))).(......).)).))))))..))))..)...... (-36.02 = -36.46 + 0.44)


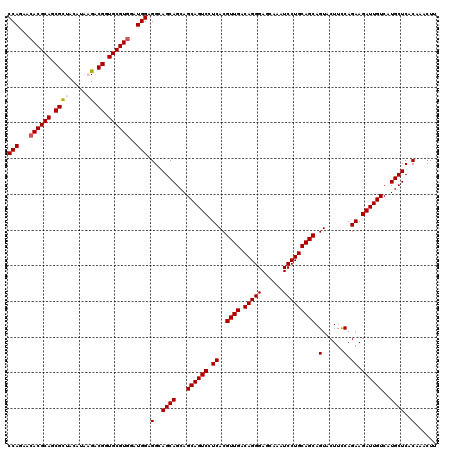
| Location | 17,128,692 – 17,128,812 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -40.76 |
| Consensus MFE | -31.80 |
| Energy contribution | -32.24 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17128692 120 - 20766785 CACUGCCCAAUACAAUCUUCGAGUUCUGGCUUAUGAUAUACCAGAACACGCAGCGCUACAUAAGACGGUGCGUGGAUGGAGGCAGCAGCAGCAGUCCUCACGUUGACAGGGAGCAAAUCC ...(((((..............(((((((...........)))))))..(((.((((.....)).)).)))((.(((((((((.((....)).).)))).)))).))..)).)))..... ( -36.10) >DroSec_CAF1 64585 120 - 1 CGUUGCCCAAUACAAUCUUCGAGUUCUGGCUUAUGAUCUACCAGAACACGCAGCGCUACAUAAGACGGUGCGUGGAUGGGGGCAGCAGCAGCAGUCCUCACGUUGACAGGGAGCAAAUCC ..((((((....((((.(((..(((((((...........)))))))(((((.((((.....)).)).))))))))(((((((.((....)).))))))).))))....)).)))).... ( -39.80) >DroSim_CAF1 55430 120 - 1 CGUUGCCCAAUACAAUCUUCGAGUUCUGGCUCAUGAUCUACCAGAACACGCAGCGCUACAUAAGACGGUGCGUGGAUGGGGGCAGCAGCAGCAGUCCUCACGUUGACAGGGAGCAAAUCC .(((((((............((((....))))........(((...((((((.((((.....)).)).))))))..))))))))))....((..((((..........))))))...... ( -40.40) >DroEre_CAF1 70113 120 - 1 CGUUGCCCAAUACUAUCUUCGAGUUCUGGCUAAUGAUCUACCAGAACACGCAGCGUUACAUUCGACGCUGCGUUGAUGGUGGAAGCAGCAGCAGUCCUCACGUUGACAGGGAGCAAAUCC .(((((....(((((((.....(((((((.((......)))))))))((((((((((......)))))))))).)))))))...))))).((..((((..........))))))...... ( -44.20) >DroYak_CAF1 72007 120 - 1 CGCUGCCCAAUACGAUCUUCGAGUUCUGGCUAAUGAUCUACCAAAACACGCAGCGCUACAUACGUCGCUGCGUGGAUGGAGGAAGCAGCAGCAGUCCUCACGUUGACAGGGAGCAAAUCC .(((.(((.....((((....(((....)))...))))........((((((((((.......).)))))))))(((((((((.((....))..))))).))))....))))))...... ( -43.30) >consensus CGUUGCCCAAUACAAUCUUCGAGUUCUGGCUAAUGAUCUACCAGAACACGCAGCGCUACAUAAGACGGUGCGUGGAUGGAGGCAGCAGCAGCAGUCCUCACGUUGACAGGGAGCAAAUCC .(((.(((..............(((((((...........)))))))..(((.((((.....)).)).)))((.((((((((..((....))...)))).)))).)).))))))...... (-31.80 = -32.24 + 0.44)

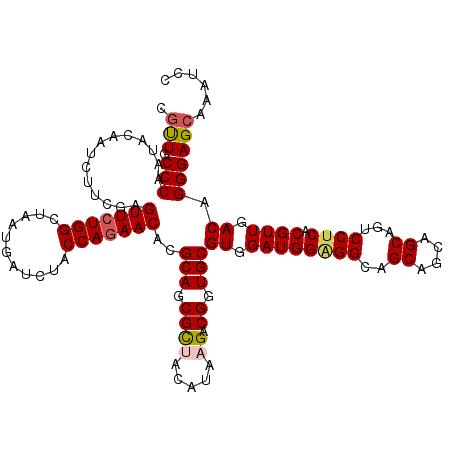
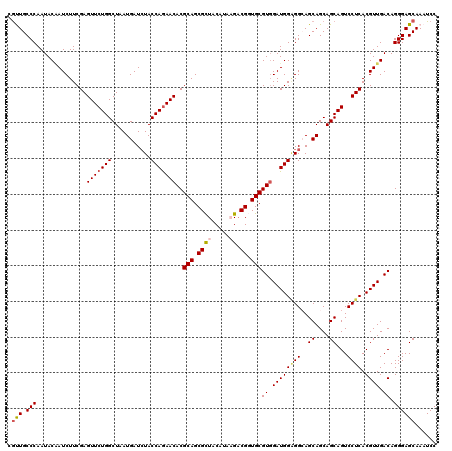
| Location | 17,128,732 – 17,128,852 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -43.32 |
| Consensus MFE | -37.78 |
| Energy contribution | -38.22 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17128732 120 - 20766785 CCCGACUACGUGUCGAAGUGCUAUGUGGCCACGCAGGGUCCACUGCCCAAUACAAUCUUCGAGUUCUGGCUUAUGAUAUACCAGAACACGCAGCGCUACAUAAGACGGUGCGUGGAUGGA .(((.(((((..(((...(..((((((((...((.((((.....))))..............(((((((...........)))))))..))...)))))))).).)))..))))).))). ( -42.50) >DroSec_CAF1 64625 120 - 1 CCCGACUACGUGUCGAAGUGCUAUGUGGCCACGCAGGGUCCGUUGCCCAAUACAAUCUUCGAGUUCUGGCUUAUGAUCUACCAGAACACGCAGCGCUACAUAAGACGGUGCGUGGAUGGG ((((.(((((..(((...(..((((((((...((.((((.....))))..............(((((((...........)))))))..))...)))))))).).)))..))))).)))) ( -44.40) >DroSim_CAF1 55470 120 - 1 CCCGACUACGUGUCGAAGUGCUAUGUGGCCACGCAGGGUCCGUUGCCCAAUACAAUCUUCGAGUUCUGGCUCAUGAUCUACCAGAACACGCAGCGCUACAUAAGACGGUGCGUGGAUGGG ((((.(((((..(((...(..((((((((...((.((((.....))))..............(((((((...........)))))))..))...)))))))).).)))..))))).)))) ( -44.40) >DroEre_CAF1 70153 120 - 1 CCCGACUACGUGUCGAAGUGCUAUGUGGCCACGCAGGGUCCGUUGCCCAAUACUAUCUUCGAGUUCUGGCUAAUGAUCUACCAGAACACGCAGCGUUACAUUCGACGCUGCGUUGAUGGU ..((((.....))))..(.((.(((.((((......))))))).)).)...((((((.....(((((((.((......)))))))))((((((((((......)))))))))).)))))) ( -45.20) >DroYak_CAF1 72047 120 - 1 CCCGACUACGUGUCGAAGUGCUAUGUGGCCACGCAGGGUCCGCUGCCCAAUACGAUCUUCGAGUUCUGGCUAAUGAUCUACCAAAACACGCAGCGCUACAUACGUCGCUGCGUGGAUGGA ..((((.....))))..(.((...((((((......)).)))).)).).....((((....(((....)))...))))..(((...((((((((((.......).)))))))))..))). ( -40.10) >consensus CCCGACUACGUGUCGAAGUGCUAUGUGGCCACGCAGGGUCCGUUGCCCAAUACAAUCUUCGAGUUCUGGCUAAUGAUCUACCAGAACACGCAGCGCUACAUAAGACGGUGCGUGGAUGGA .(((.((((((..((......((((((((...((.((((.....))))..............(((((((...........)))))))..))...))))))))...))..)))))).))). (-37.78 = -38.22 + 0.44)

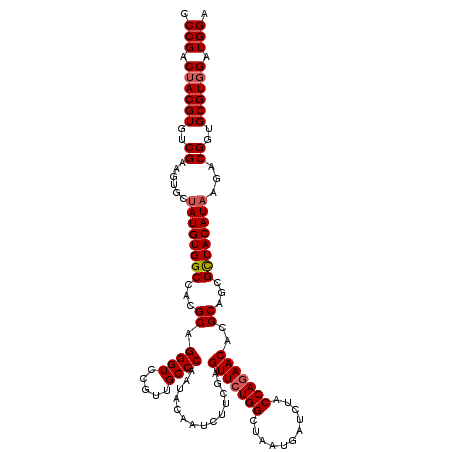
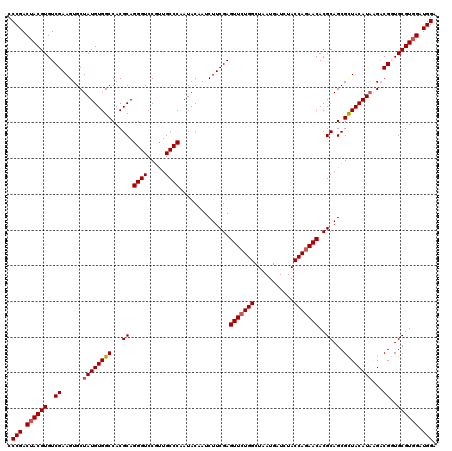
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:36 2006