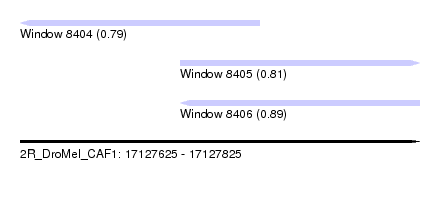
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,127,625 – 17,127,825 |
| Length | 200 |
| Max. P | 0.890147 |

| Location | 17,127,625 – 17,127,745 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -42.33 |
| Consensus MFE | -39.24 |
| Energy contribution | -39.52 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17127625 120 - 20766785 GUUUGCAAUCUUCGCCUGCAGCGGGGCGGGAUGGUCCAGAACUCGGAGCAGUAUGAGCUCAUCCAUCGGGCCAUUUGCCUCUACCUGAAACGCACACUGGCGUUGAGGCGAUUUUAAAAG ...........((((((.(((.(((((((.(((((((.((.....((((.......)))).....))))))))))))))))...))).(((((......))))).))))))......... ( -44.00) >DroSec_CAF1 63518 120 - 1 GUUUGUAAUCUUCGCCUGCAGCGGGGCGGGAUGGUCCAGAACUCGGAGCAGUAUGAGCUCAUCCAUCGGGCCAUUUGCCUCUAUCUGAAACGCACACUGGCGUUGAGGCGAUUUUAAAAG .....(((...((((((.(((.(((((((.(((((((.((.....((((.......)))).....))))))))))))))))...))).(((((......))))).))))))..))).... ( -44.10) >DroSim_CAF1 54363 120 - 1 GUUUGUAAUCUUCGCCUGCAGCGGGGCGGGAUGGUCCAAAACUCGGAGCAGUAUGAGCUCAUCCAUCGGGCCAUUUGCCUCUAUCUGAAACGCACACUGGCGUUGAGGCGAUUUUAAAAG .....(((...((((((.(((.(((((((.(((((((........((((.......)))).......))))))))))))))...))).(((((......))))).))))))..))).... ( -42.86) >DroEre_CAF1 69046 120 - 1 GUUUGCAAUCUUCGCUUGCAGCGGGGCGGGAUGGUCCAGAACUCGGAGCAGUAUGAGCUAAUCCAUCGGGCCAUUUGCCUCUAUCUGAAACGGACACUGGCGUUGAGGCGAUUUUAAAAG ...........((((((.(((((((((((.(((((((.(....)(((..(((....)))..)))...))))))))))))))..((((...)))).......))))))))))......... ( -39.40) >DroYak_CAF1 70940 120 - 1 GUUUGCAAUCUUCGCCUGCAGCGGGGCGGUAUGGUCCAGAACUCGGAGCAGUAUGAGCUAAUACACCGGGCCAUUUGCCUCUAUCUGAAGCGCACACUCGCGUUGAGGCGAUUUUAAAAG ...........((((((.(((((((((((.(((((((.(....)((....((((......)))).))))))))))))))))........(((......)))))))))))))......... ( -41.30) >consensus GUUUGCAAUCUUCGCCUGCAGCGGGGCGGGAUGGUCCAGAACUCGGAGCAGUAUGAGCUCAUCCAUCGGGCCAUUUGCCUCUAUCUGAAACGCACACUGGCGUUGAGGCGAUUUUAAAAG ...........((((((.(((.(((((((.(((((((........((((.......)))).......))))))))))))))...))).(((((......))))).))))))......... (-39.24 = -39.52 + 0.28)

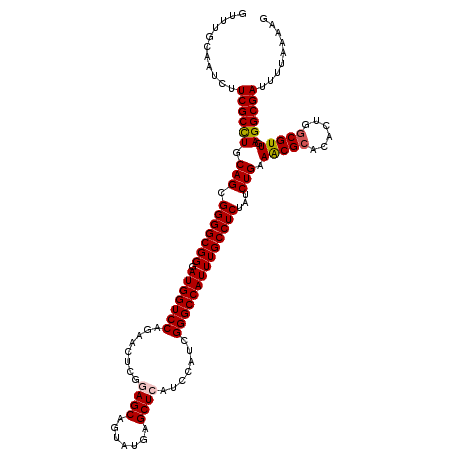
| Location | 17,127,705 – 17,127,825 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -46.30 |
| Consensus MFE | -37.54 |
| Energy contribution | -38.18 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17127705 120 + 20766785 UCUGGACCAUCCCGCCCCGCUGCAGGCGAAGAUUGCAAACGAUGCCCAGGACGUCCACGAAAAUGUCCGGCAUCUUGGGCAGACGGUCAAAGGAUGGCGGCGUCUUAACCGCCUCCGCAU ....((((.((..((((...(((((.......)))))...((((((..((((((........))))))))))))..)))).)).))))...(((.(((((........)))))))).... ( -49.10) >DroSec_CAF1 63598 120 + 1 UCUGGACCAUCCCGCCCCGCUGCAGGCGAAGAUUACAAACGAUGCCCAGGACGUCCACGAAAAUGUCCGGCAUCUUGGGCAGACGGUCAAAGGAUGGCGGCGUCUUAACCGCCCCUGCCU ....((((.((..(((((((.....)))............((((((..((((((........))))))))))))..)))).)).))))..(((..(((((........)))))....))) ( -46.30) >DroSim_CAF1 54443 120 + 1 UUUGGACCAUCCCGCCCCGCUGCAGGCGAAGAUUACAAACGAUGCCCAGGACGUCCACGAAAAUGUCCGGCAUCUUGGGCAGACGGUCAAAGGAUGGCGGCGUCUUAACCGCCCCUGCCU ....((((.((..(((((((.....)))............((((((..((((((........))))))))))))..)))).)).))))..(((..(((((........)))))....))) ( -46.30) >DroEre_CAF1 69126 120 + 1 UCUGGACCAUCCCGCCCCGCUGCAAGCGAAGAUUGCAAACGAUGCCCAAGACGUCCACGAAAAUGUCCGGCAUCUUAGGCAGACGGUCGAAGGAUGCGGGCGCCUUAGCCGCCUCCGUCU ....((((.((..(((....(((((.......)))))...((((((...(((((........))))).))))))...))).)).))))((.(((.((((.(......))))).))).)). ( -42.40) >DroYak_CAF1 71020 120 + 1 UCUGGACCAUACCGCCCCGCUGCAGGCGAAGAUUGCAAACGAUGCCCAGGACGUCCACGAAAAUGUCCGGCAUCUUAGGCAGACGGUCGAAUGAGGGGGGCGUCGUAACUGCCUCCGUCU ....((((....((((........))))..(.((((....((((((..((((((........))))))))))))....)))).)))))....((.(((((((.......))))))).)). ( -47.40) >consensus UCUGGACCAUCCCGCCCCGCUGCAGGCGAAGAUUGCAAACGAUGCCCAGGACGUCCACGAAAAUGUCCGGCAUCUUGGGCAGACGGUCAAAGGAUGGCGGCGUCUUAACCGCCUCCGCCU (((.((((.((..(((((((.....)))............((((((..((((((........))))))))))))..)))).)).))))...))).((.((((.......)))).)).... (-37.54 = -38.18 + 0.64)

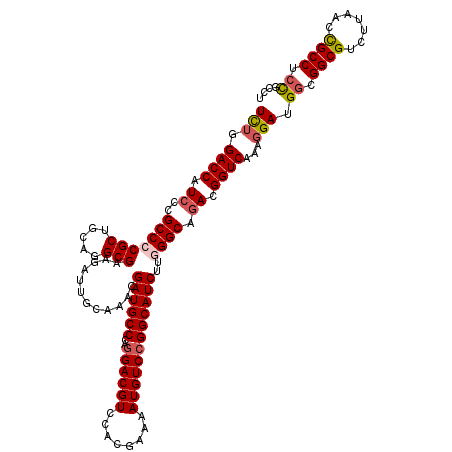
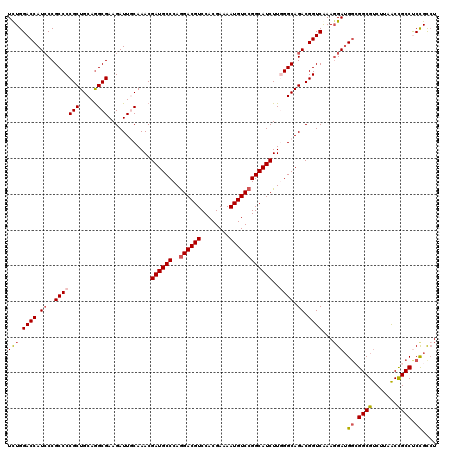
| Location | 17,127,705 – 17,127,825 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -50.54 |
| Consensus MFE | -45.30 |
| Energy contribution | -45.54 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
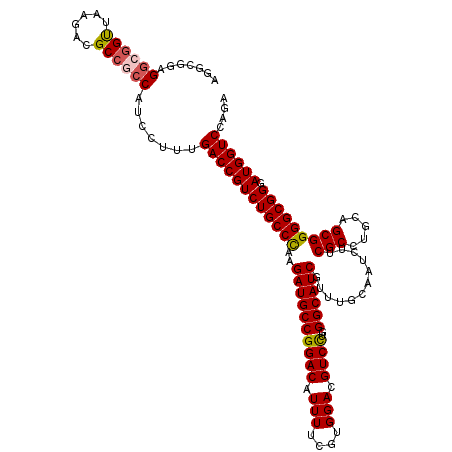
>2R_DroMel_CAF1 17127705 120 - 20766785 AUGCGGAGGCGGUUAAGACGCCGCCAUCCUUUGACCGUCUGCCCAAGAUGCCGGACAUUUUCGUGGACGUCCUGGGCAUCGUUUGCAAUCUUCGCCUGCAGCGGGGCGGGAUGGUCCAGA ....(((((((((......)))))).)))...((((((((((((..((((((((((.(((....))).))))..))))))(((.(((.........)))))).)))).)))))))).... ( -54.00) >DroSec_CAF1 63598 120 - 1 AGGCAGGGGCGGUUAAGACGCCGCCAUCCUUUGACCGUCUGCCCAAGAUGCCGGACAUUUUCGUGGACGUCCUGGGCAUCGUUUGUAAUCUUCGCCUGCAGCGGGGCGGGAUGGUCCAGA (((....((((((......))))))..)))..((((((((((((..((((((((((.(((....))).))))..))))))............(((.....))))))).)))))))).... ( -51.20) >DroSim_CAF1 54443 120 - 1 AGGCAGGGGCGGUUAAGACGCCGCCAUCCUUUGACCGUCUGCCCAAGAUGCCGGACAUUUUCGUGGACGUCCUGGGCAUCGUUUGUAAUCUUCGCCUGCAGCGGGGCGGGAUGGUCCAAA (((....((((((......))))))..)))..((((((((((((..((((((((((.(((....))).))))..))))))............(((.....))))))).)))))))).... ( -51.20) >DroEre_CAF1 69126 120 - 1 AGACGGAGGCGGCUAAGGCGCCCGCAUCCUUCGACCGUCUGCCUAAGAUGCCGGACAUUUUCGUGGACGUCUUGGGCAUCGUUUGCAAUCUUCGCUUGCAGCGGGGCGGGAUGGUCCAGA .((.(((.(((((......).)))).))).))(((((((((((((((((((((((.....)).))).)))))))))).(((((.((((.......)))))))))....)))))))).... ( -50.80) >DroYak_CAF1 71020 120 - 1 AGACGGAGGCAGUUACGACGCCCCCCUCAUUCGACCGUCUGCCUAAGAUGCCGGACAUUUUCGUGGACGUCCUGGGCAUCGUUUGCAAUCUUCGCCUGCAGCGGGGCGGUAUGGUCCAGA .((.((.(((.((....))))).)).))....((((((((((((..((((((((((.(((....))).))))..))))))(((.(((.........)))))).)))))).)))))).... ( -45.50) >consensus AGGCGGAGGCGGUUAAGACGCCGCCAUCCUUUGACCGUCUGCCCAAGAUGCCGGACAUUUUCGUGGACGUCCUGGGCAUCGUUUGCAAUCUUCGCCUGCAGCGGGGCGGGAUGGUCCAGA .......((((((......)))))).......((((((((((((..((((((((((.(((....))).))))..))))))............(((.....))))))))).)))))).... (-45.30 = -45.54 + 0.24)

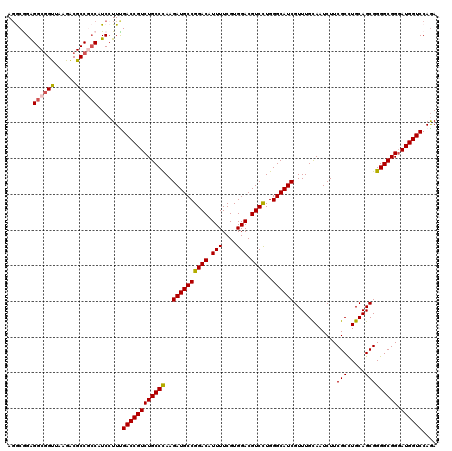
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:29 2006