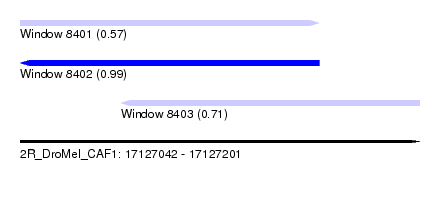
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,127,042 – 17,127,201 |
| Length | 159 |
| Max. P | 0.990630 |

| Location | 17,127,042 – 17,127,161 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.89 |
| Mean single sequence MFE | -40.88 |
| Consensus MFE | -34.44 |
| Energy contribution | -35.00 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17127042 119 + 20766785 UCCGGCGGUCGAGCUGUCCGCUUGGGAUUGGAGAAAUUGGUGUU-UAGUCAGAAGGACCCAGGGAAGGAUCCAACUUGAGAAAGGUUUGAGCUUUCUCCGCCUCCAAGCAGGAUGCACAA ((((((((.....))))).(((((((..((((((((.....(((-((.......(((.((......)).)))(((((.....))))))))))))))))))..))))))).)))....... ( -38.90) >DroSec_CAF1 62936 119 + 1 UCCGGCGGUCGAGCUGCCCGCUUGGGAUUGGAGAAAUUGGUGUU-UAGUCAGAAGGACCCAGGGUAGGAUCCAACUUGAGAAAGGUUUGAGCUUUCUCCGCCUCCAAGCAGGAUGCACAA ((((((((.....))))).(((((((..((((((((.....(((-((.......(((.((......)).)))(((((.....))))))))))))))))))..))))))).)))....... ( -41.60) >DroSim_CAF1 53781 119 + 1 UCCGGCGGUCGAGCUGCCCGCUUGGGAUUGGAGAAAUUGGUGUU-UAGUCAGAAGGACCCAGGGUAGGAUCCGACUUGAGAAAGGUUUGAGCUUUCUCCGCCUCCAAGCAGGAUGCACAA ((((((((.....))))).(((((((..((((((((.....(((-((.......(((.((......)).)))(((((.....))))))))))))))))))..))))))).)))....... ( -42.00) >DroEre_CAF1 68462 120 + 1 UCCGGCGGUCGAGCUGUCAGCUUGGGAUUGGAGAAAUUGGUGUUUUAGUCAGAAGGACCCCGCGAAGGGUCCAACUUGAAAAAGGUUUGAGCUUUCUCCGCACCCAAACAGGAUGCACAA ((((((((.....))))).(.(((((..((((((((((((....))))(((((.((((((......))))))..(((....))).)))))..))))))))..))))).).)))....... ( -41.10) >DroYak_CAF1 70375 114 + 1 UCCGGCGGUCGAGCUGUCAGCUCGGGAUUGGAGAAAUUGGUGUUUUAGUCAGAAGGACCCUGGGAAGGGUCCAACUUGAAAAAGGUUUGAGCUUUCUCCGUACCCAAAC------CCAAA ....(.(((.((((.....))))(((..((((((((((((....))))(((((.(((((((....)))))))..(((....))).)))))..))))))))..)))..))------))... ( -40.80) >consensus UCCGGCGGUCGAGCUGUCCGCUUGGGAUUGGAGAAAUUGGUGUU_UAGUCAGAAGGACCCAGGGAAGGAUCCAACUUGAGAAAGGUUUGAGCUUUCUCCGCCUCCAAGCAGGAUGCACAA ((((((((.....))))).(((((((..((((((((..................(((.((......)).)))(((((.....))))).....))))))))..))))))).)))....... (-34.44 = -35.00 + 0.56)

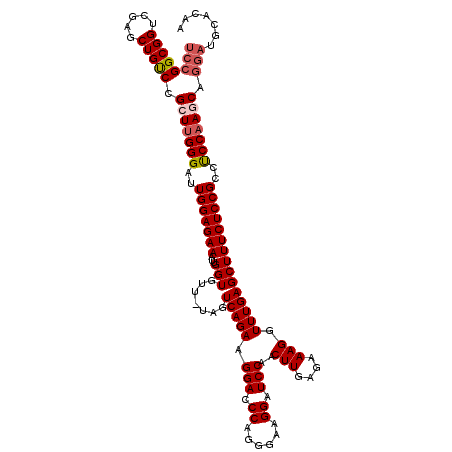
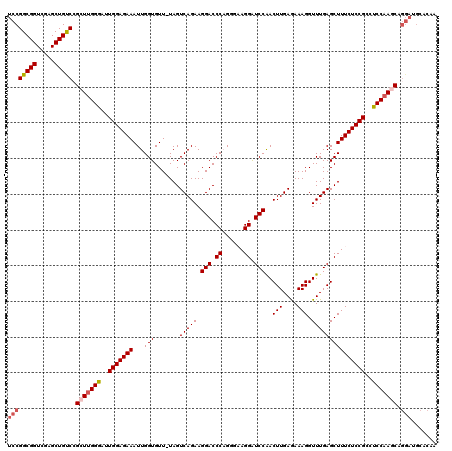
| Location | 17,127,042 – 17,127,161 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.89 |
| Mean single sequence MFE | -37.84 |
| Consensus MFE | -32.41 |
| Energy contribution | -32.77 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17127042 119 - 20766785 UUGUGCAUCCUGCUUGGAGGCGGAGAAAGCUCAAACCUUUCUCAAGUUGGAUCCUUCCCUGGGUCCUUCUGACUA-AACACCAAUUUCUCCAAUCCCAAGCGGACAGCUCGACCGCCGGA (((.((...((((((((.((.(((((((.............(((.(..((((((......))))))..))))...-........)))))))..))))))))))...)).)))........ ( -35.55) >DroSec_CAF1 62936 119 - 1 UUGUGCAUCCUGCUUGGAGGCGGAGAAAGCUCAAACCUUUCUCAAGUUGGAUCCUACCCUGGGUCCUUCUGACUA-AACACCAAUUUCUCCAAUCCCAAGCGGGCAGCUCGACCGCCGGA (((.((..(((((((((.((.(((((((.............(((.(..((((((......))))))..))))...-........)))))))..)))))))))))..)).)))........ ( -39.05) >DroSim_CAF1 53781 119 - 1 UUGUGCAUCCUGCUUGGAGGCGGAGAAAGCUCAAACCUUUCUCAAGUCGGAUCCUACCCUGGGUCCUUCUGACUA-AACACCAAUUUCUCCAAUCCCAAGCGGGCAGCUCGACCGCCGGA (((.((..(((((((((.(((.(((((((.......)))))))..)))((((((......)))))).........-...................)))))))))..)).)))........ ( -39.80) >DroEre_CAF1 68462 120 - 1 UUGUGCAUCCUGUUUGGGUGCGGAGAAAGCUCAAACCUUUUUCAAGUUGGACCCUUCGCGGGGUCCUUCUGACUAAAACACCAAUUUCUCCAAUCCCAAGCUGACAGCUCGACCGCCGGA (((.((.((..(((((((...(((((((.............(((.(..(((((((....)))))))..))))............)))))))...))))))).))..)).)))........ ( -37.01) >DroYak_CAF1 70375 114 - 1 UUUGG------GUUUGGGUACGGAGAAAGCUCAAACCUUUUUCAAGUUGGACCCUUCCCAGGGUCCUUCUGACUAAAACACCAAUUUCUCCAAUCCCGAGCUGACAGCUCGACCGCCGGA ....(------(((.(((...(((((((.............(((.(..(((((((....)))))))..))))............)))))))...)))((((.....))))))))...... ( -37.81) >consensus UUGUGCAUCCUGCUUGGAGGCGGAGAAAGCUCAAACCUUUCUCAAGUUGGAUCCUUCCCUGGGUCCUUCUGACUA_AACACCAAUUUCUCCAAUCCCAAGCGGACAGCUCGACCGCCGGA (((.((...((((((((....(((((((.............(((.(..((((((......))))))..))))............)))))))....))))))))...)).)))........ (-32.41 = -32.77 + 0.36)

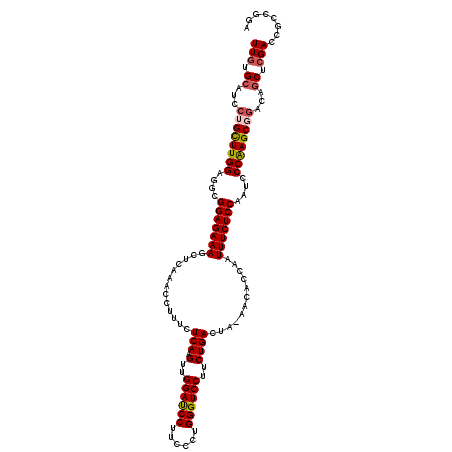
| Location | 17,127,082 – 17,127,201 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.22 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -33.04 |
| Energy contribution | -32.12 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
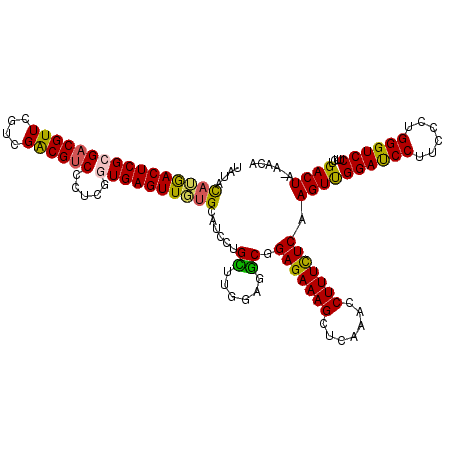
>2R_DroMel_CAF1 17127082 119 - 20766785 UAUAUAUGACUCGCGACGUUCGUCGACGCCCCUCGGUGAGUUGUGCAUCCUGCUUGGAGGCGGAGAAAGCUCAAACCUUUCUCAAGUUGGAUCCUUCCCUGGGUCCUUCUGACUA-AACA .....(..(((((((((....)))).)(((....)))))))..)((.(((.....))).)).(((((((.......))))))).((..((((((......))))))..)).....-.... ( -37.20) >DroSec_CAF1 62976 119 - 1 UAUACAUGACUCGCGACGUUCGUCGACGUCCCUUGGUGAGUUGUGCAUCCUGCUUGGAGGCGGAGAAAGCUCAAACCUUUCUCAAGUUGGAUCCUACCCUGGGUCCUUCUGACUA-AACA .....(..((((((((((((....)))))).....))))))..)((.(((.....))).)).(((((((.......))))))).((..((((((......))))))..)).....-.... ( -38.50) >DroSim_CAF1 53821 119 - 1 UAUACAUGACUCGCGACGUUCGUCGACGUCCCUCGGUGAGUUGUGCAUCCUGCUUGGAGGCGGAGAAAGCUCAAACCUUUCUCAAGUCGGAUCCUACCCUGGGUCCUUCUGACUA-AACA .....(..((((((((((((....)))))).....))))))..)((.(((.....))).)).(((((((.......))))))).((((((((((......))))))....)))).-.... ( -40.10) >DroEre_CAF1 68502 120 - 1 UAUACAAAACUCGAGACGUUCGUCGGCGUCCCUCGGUGAGUUGUGCAUCCUGUUUGGGUGCGGAGAAAGCUCAAACCUUUUUCAAGUUGGACCCUUCGCGGGGUCCUUCUGACUAAAACA ...((((.((.((((((((......)))))..)))))...))))((((((.....)))))).(((((((.......))))))).((..(((((((....)))))))..)).......... ( -39.20) >DroYak_CAF1 70415 114 - 1 UAUACAAAACUCGAGACGUUCGUCGGCGUCCCUCGGUGAGUUUGG------GUUUGGGUACGGAGAAAGCUCAAACCUUUUUCAAGUUGGACCCUUCCCAGGGUCCUUCUGACUAAAACA ..........(((((((((......)))))..))))((((...((------((((((((.........))))))))))..))))((..(((((((....)))))))..)).......... ( -36.00) >consensus UAUACAUGACUCGCGACGUUCGUCGACGUCCCUCGGUGAGUUGUGCAUCCUGCUUGGAGGCGGAGAAAGCUCAAACCUUUCUCAAGUUGGAUCCUUCCCUGGGUCCUUCUGACUA_AACA ....((((((((((((((((....)))))).....))))))))))......((......)).(((((((.......))))))).((((((((((......))))))....))))...... (-33.04 = -32.12 + -0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:27 2006